class: center, middle, inverse, title-slide .title[ # Single-cell RNA sequencing ~ Session 2<br /> <html><br /> <br /> <hr color='#EB811B' size=1px width=796px><br /> </html> ] .author[ ### Rockefeller University, Bioinformatics Resource Centre ] .date[ ### <a href="http://rockefelleruniversity.github.io/scRNA-seq/" class="uri">http://rockefelleruniversity.github.io/scRNA-seq/</a> ] --- ## Overview In this course we are going to introduce basic analysis for single-cell RNAseq, with a specific focus on the *10X system*. The course is divided into multiple sessions. In this second session, we will introduce Seurat and Bioconductor methodologies for basic scRNA-seq QC, filtering and initial analysis of single samples. --- ## The data For these sessions we are going to make use of two datasets. The first set will be from the recent paper [**Enteroendocrine cell lineages that differentially control feeding and gut motility**](https://elifesciences.org/articles/78512). This contains scRNA data from either Neurod1 and Neurog3 expressing enteroendocrine cells. The second dataset is the classic example from PBMC cells. --- ## Seurat and Bioconductor The Seurat R package and associated packages offer an R based and well established methodology for the analysis of single cell RNA-seq, ATAC-seq and many other single-cell sequencing methodologies. [Seurat page](https://satijalab.org/seurat/) Bioconductor has a set of interrelated and highly connected software packages for single cell RNAseq as well as integrating with the rest of the Bioconductor software ecosystem. [OSCA book](https://bioconductor.org/books/3.18/OSCA/) In this session we will run through basic analyses using both of the methodologies to show both their equvalence in some parts of the analysis as well as their distinct functionalities in others. In practice, you may select methods from either softwares and convert between the two as required. --- # The Example Data For this session we will need the filtered and raw expression data for the Neurod1 datasets. The filtered matrix can be found [here](https://rubioinformatics.s3.amazonaws.com/scRNA_graduate/SRR_NeuroD1/outs/NeuroD1_filtered_feature_bc_matrix.h5) The raw matrix can be found here [here](https://rubioinformatics.s3.amazonaws.com/scRNA_graduate/SRR_NeuroD1/outs/raw_feature_bc_matrix.h5) --- class: inverse, center, middle # Bioconductor methods <html><div style='float:left'></div><hr color='#EB811B' size=1px width=720px></html> --- --- # Data import (Bioconductor) First we need to load the DropletUtils package to read and handle our Droplet 10X data. ``` r library(DropletUtils) ``` ``` ## Warning: package 'DropletUtils' was built under R version 4.4.1 ``` ``` ## Loading required package: SingleCellExperiment ``` ``` ## Warning: package 'SingleCellExperiment' was built under R version 4.4.1 ``` ``` ## Loading required package: SummarizedExperiment ``` ``` ## Warning: package 'SummarizedExperiment' was built under R version 4.4.1 ``` ``` ## Loading required package: MatrixGenerics ``` ``` ## Warning: package 'MatrixGenerics' was built under R version 4.4.1 ``` ``` ## Loading required package: matrixStats ``` ``` ## ## Attaching package: 'MatrixGenerics' ``` ``` ## The following objects are masked from 'package:matrixStats': ## ## colAlls, colAnyNAs, colAnys, colAvgsPerRowSet, colCollapse, ## colCounts, colCummaxs, colCummins, colCumprods, colCumsums, ## colDiffs, colIQRDiffs, colIQRs, colLogSumExps, colMadDiffs, ## colMads, colMaxs, colMeans2, colMedians, colMins, colOrderStats, ## colProds, colQuantiles, colRanges, colRanks, colSdDiffs, colSds, ## colSums2, colTabulates, colVarDiffs, colVars, colWeightedMads, ## colWeightedMeans, colWeightedMedians, colWeightedSds, ## colWeightedVars, rowAlls, rowAnyNAs, rowAnys, rowAvgsPerColSet, ## rowCollapse, rowCounts, rowCummaxs, rowCummins, rowCumprods, ## rowCumsums, rowDiffs, rowIQRDiffs, rowIQRs, rowLogSumExps, ## rowMadDiffs, rowMads, rowMaxs, rowMeans2, rowMedians, rowMins, ## rowOrderStats, rowProds, rowQuantiles, rowRanges, rowRanks, ## rowSdDiffs, rowSds, rowSums2, rowTabulates, rowVarDiffs, rowVars, ## rowWeightedMads, rowWeightedMeans, rowWeightedMedians, ## rowWeightedSds, rowWeightedVars ``` ``` ## Loading required package: GenomicRanges ``` ``` ## Warning: package 'GenomicRanges' was built under R version 4.4.1 ``` ``` ## Loading required package: GenomeInfoDb ``` ``` ## Warning: package 'GenomeInfoDb' was built under R version 4.4.1 ``` ``` ## ## Attaching package: 'GenomicRanges' ``` ``` ## The following object is masked from 'package:magrittr': ## ## subtract ``` ``` ## Loading required package: Biobase ``` ``` ## Warning: package 'Biobase' was built under R version 4.4.1 ``` ``` ## Welcome to Bioconductor ## ## Vignettes contain introductory material; view with ## 'browseVignettes()'. To cite Bioconductor, see ## 'citation("Biobase")', and for packages 'citation("pkgname")'. ``` ``` ## ## Attaching package: 'Biobase' ``` ``` ## The following object is masked from 'package:MatrixGenerics': ## ## rowMedians ``` ``` ## The following objects are masked from 'package:matrixStats': ## ## anyMissing, rowMedians ``` --- # Read10x We can then read in the filtered matrix containing data on droplets marked as cells by CellRanger. We set the row.names argument for easier interpretation. ``` r h5file <- "https://rubioinformatics.s3.amazonaws.com/scRNA_graduate/SRR_NeuroD1/outs/filtered_feature_bc_matrix.h5" local_h5file <- basename(h5file) download.file(h5file, local_h5file) sce.NeuroD1_filtered <- read10xCounts(local_h5file, col.names = TRUE, row.names = "symbol") class(sce.NeuroD1_filtered) ``` ``` ## [1] "SingleCellExperiment" ## attr(,"package") ## [1] "SingleCellExperiment" ``` --- ## SingleCellExperiment Object The created SingleCellExperiment Object is much like our SummarizedExperiment object ``` r sce.NeuroD1_filtered ``` ``` ## class: SingleCellExperiment ## dim: 39905 3468 ## metadata(1): Samples ## assays(1): counts ## rownames(39905): Gm26206 Xkr4 ... MT-TrnT MT-TrnP ## rowData names(3): ID Symbol Type ## colnames(3468): AAACCCACACAATGTC-1 AAACCCACACCACTGG-1 ... ## TTTGTTGTCTCTTAAC-1 TTTGTTGTCTGGACCG-1 ## colData names(2): Sample Barcode ## reducedDimNames(0): ## mainExpName: NULL ## altExpNames(0): ``` --- ## SingleCellExperiment Object The *colnames()* and *rownames()* functions can be used to access column names (cell-barcodes) and row names (gene identifiers) ``` r # cell information colnames(sce.NeuroD1_filtered)[1:2] ``` ``` ## [1] "AAACCCACACAATGTC-1" "AAACCCACACCACTGG-1" ``` ``` r # gene information rownames(sce.NeuroD1_filtered)[1:2] ``` ``` ## [1] "Gm26206" "Xkr4" ``` --- ## SingleCellExperiment Object The *colData()* and *rowData()* functions can be used to access experiment metadata. ``` r # cell information colData(sce.NeuroD1_filtered)[1:2, ] ``` ``` ## DataFrame with 2 rows and 2 columns ## Sample Barcode ## <character> <character> ## AAACCCACACAATGTC-1 filtered_feature_bc_.. AAACCCACACAATGTC-1 ## AAACCCACACCACTGG-1 filtered_feature_bc_.. AAACCCACACCACTGG-1 ``` ``` r # gene information rowData(sce.NeuroD1_filtered)[1:2, ] ``` ``` ## DataFrame with 2 rows and 3 columns ## ID Symbol Type ## <character> <character> <character> ## Gm26206 Gm26206 Gm26206 Gene Expression ## Xkr4 Xkr4 Xkr4 Gene Expression ``` --- ## SingleCellExperiment Object A reducedDim and reducedDimNames slots at present remains unfilled. The metadata slot contains the Sample names. Here all these cells came from single sample ``` r # cell information reducedDimNames(sce.NeuroD1_filtered) ``` ``` ## character(0) ``` ``` r # gene information metadata(sce.NeuroD1_filtered) ``` ``` ## $Samples ## [1] "filtered_feature_bc_matrix.h5" ``` --- ## The knee plot One of the first plots we may want to recreate from the QC is the knee plot. To do this we will need the unfiltered matrix too. The unfiltered SingleCellExperiment contains over a million droplets. ``` r h5file <- "https://rubioinformatics.s3.amazonaws.com/scRNA_graduate/SRR_NeuroD1/outs/raw_feature_bc_matrix.h5" local_h5file <- "NeuroD1_raw_feature_bc_matrix.h5" download.file(h5file, local_h5file) sce.NeuroD1_unfiltered <- read10xCounts(local_h5file, col.names = TRUE) sce.NeuroD1_unfiltered ``` ``` ## class: SingleCellExperiment ## dim: 39905 1244317 ## metadata(1): Samples ## assays(1): counts ## rownames(39905): Gm26206 Xkr4 ... MT-TrnT MT-TrnP ## rowData names(3): ID Symbol Type ## colnames(1244317): AAACCCAAGAAACACT-1 AAACCCAAGAAACCAT-1 ... ## TTTGTTGTCTTTGCTA-1 TTTGTTGTCTTTGTCG-1 ## colData names(2): Sample Barcode ## reducedDimNames(0): ## mainExpName: NULL ## altExpNames(0): ``` --- ## Barcode ranks We can use the *barcodeRanks* function to create a table of barcode ranks and total reads per droplet. ``` r bcrank <- barcodeRanks(counts(sce.NeuroD1_unfiltered)) bcrank ``` ``` ## DataFrame with 1244317 rows and 3 columns ## rank total fitted ## <numeric> <integer> <numeric> ## AAACCCAAGAAACACT-1 45958 155 NA ## AAACCCAAGAAACCAT-1 102639 4 NA ## AAACCCAAGAAACCCG-1 629672 1 NA ## AAACCCAAGAAACCTG-1 629672 1 NA ## AAACCCAAGAAAGAAC-1 1096187 0 NA ## ... ... ... ... ## TTTGTTGTCTTTGATC-1 36930.5 167 NA ## TTTGTTGTCTTTGCAT-1 629672.5 1 NA ## TTTGTTGTCTTTGCGC-1 629672.5 1 NA ## TTTGTTGTCTTTGCTA-1 629672.5 1 NA ## TTTGTTGTCTTTGTCG-1 629672.5 1 NA ``` --- ## Retrieve filtered barcodes We then filter for droplets with the same rank for visualisation purposes. ``` r bcrank$filtered <- rownames(bcrank) %in% colnames(sce.NeuroD1_filtered) bc_plot <- as.data.frame(bcrank) bc_plot <- bc_plot[order(bc_plot$filtered, decreasing = TRUE), ] bc_plot <- bc_plot[!duplicated(bc_plot$rank), ] ``` --- ## Plotting barcode rank We can now recreate the knee plot from CellRanger. ``` r require(ggplot2) ``` ``` ## Loading required package: ggplot2 ``` ``` r ggplot(bc_plot, aes(x = rank, y = total, colour = filtered, alpha = 0.001)) + geom_point() + scale_y_log10() + scale_x_log10() + theme_minimal() + geom_hline(yintercept = metadata(bcrank)$inflection, colour = "darkgreen", linetype = 2) + geom_hline(yintercept = metadata(bcrank)$knee, colour = "dodgerblue", linetype = 2) ``` ``` ## Warning in scale_y_log10(): log-10 transformation introduced infinite values. ``` <!-- --> --- ## Filtering droplets using Droplet utils Although CellRanger provides a filtered Droplet matrix, you may wish to provide your own cut-off based on knee plots generated and/or use the emptyDrops method in Droplet utils to identify cell containing droplets. ``` r e.out <- emptyDrops(counts(sce.NeuroD1_unfiltered)) table(e.out) head(e.out) ``` ``` ## DataFrame with 6 rows and 5 columns ## Total LogProb PValue Limited FDR ## <integer> <numeric> <numeric> <logical> <numeric> ## AAACCCAAGAAACACT-1 118 -350.537 0.30007 FALSE 1 ## AAACCCAAGAAACCAT-1 2 NA NA NA NA ## AAACCCAAGAAACCCG-1 0 NA NA NA NA ## AAACCCAAGAAACCTG-1 0 NA NA NA NA ## AAACCCAAGAAAGAAC-1 0 NA NA NA NA ## AAACCCAAGAAAGCGA-1 1 NA NA NA NA ``` --- class: inverse, center, middle # Bioconductor - QC <html><div style='float:left'></div><hr color='#EB811B' size=1px width=720px></html> --- ## Working with CellRanger filtered matrix From this point we will use the Droplets filtered using Cellranger ``` r sce.NeuroD1_filtered ``` ``` ## class: SingleCellExperiment ## dim: 39905 3468 ## metadata(1): Samples ## assays(1): counts ## rownames(39905): Gm26206 Xkr4 ... MT-TrnT MT-TrnP ## rowData names(3): ID Symbol Type ## colnames(3468): AAACCCACACAATGTC-1 AAACCCACACCACTGG-1 ... ## TTTGTTGTCTCTTAAC-1 TTTGTTGTCTGGACCG-1 ## colData names(2): Sample Barcode ## reducedDimNames(0): ## mainExpName: NULL ## altExpNames(0): ``` --- ## Add QC metrics To assess QC we first want to identify mitochondrial and ribosomal genes. High mitochondrial gene expression within a cell is often used as a marker of dying cells. ``` r is.mito <- grepl("^MT", rowData(sce.NeuroD1_filtered)$Symbol) # is.ribo <- grepl('^Rps',rowData(sce.NeuroD1_filtered)$Symbol) table(is.mito) ``` ``` ## is.mito ## FALSE TRUE ## 39868 37 ``` ``` r # table(is.ribo) ``` --- ## Add QC metrics The scuttle package has functions for normalisation, transformation and QC. Here we us the addPerCellQCMetrics function to update our SingleCellExperiment object with QC information. ``` r library(scuttle) ``` ``` ## Warning: package 'scuttle' was built under R version 4.4.1 ``` ``` r sce.NeuroD1_filtered <- addPerCellQCMetrics(sce.NeuroD1_filtered, subsets = list(Mito = is.mito)) sce.NeuroD1_filtered ``` ``` ## class: SingleCellExperiment ## dim: 39905 3468 ## metadata(1): Samples ## assays(1): counts ## rownames(39905): Gm26206 Xkr4 ... MT-TrnT MT-TrnP ## rowData names(3): ID Symbol Type ## colnames(3468): AAACCCACACAATGTC-1 AAACCCACACCACTGG-1 ... ## TTTGTTGTCTCTTAAC-1 TTTGTTGTCTGGACCG-1 ## colData names(8): Sample Barcode ... subsets_Mito_percent total ## reducedDimNames(0): ## mainExpName: NULL ## altExpNames(0): ``` --- ## QC in colData The QC data per Droplet has been added to additional columns in the colData slot of our SingleCellExperiment. ``` r qc_df <- colData(sce.NeuroD1_filtered) head(qc_df, n = 2) ``` ``` ## DataFrame with 2 rows and 8 columns ## Sample Barcode sum ## <character> <character> <numeric> ## AAACCCACACAATGTC-1 filtered_feature_bc_.. AAACCCACACAATGTC-1 23320 ## AAACCCACACCACTGG-1 filtered_feature_bc_.. AAACCCACACCACTGG-1 1944 ## detected subsets_Mito_sum subsets_Mito_detected ## <integer> <numeric> <integer> ## AAACCCACACAATGTC-1 4776 3358 19 ## AAACCCACACCACTGG-1 176 1577 15 ## subsets_Mito_percent total ## <numeric> <numeric> ## AAACCCACACAATGTC-1 14.3997 23320 ## AAACCCACACCACTGG-1 81.1214 1944 ``` --- ## Plotting QC data We can then plot this using the plotColData function from the Scater package. First we plot the distritbution of total reads per cell. ``` r library(scater) ``` ``` ## Warning: package 'scater' was built under R version 4.4.1 ``` ``` r plotColData(sce.NeuroD1_filtered, x = "Sample", y = "sum") ``` <!-- --> --- ## Plotting QC data Then we plot the distritbution of total detected genes per cell. ``` r library(scater) plotColData(sce.NeuroD1_filtered, x = "Sample", y = "detected") ``` <!-- --> --- ## Plotting QC data Then we plot the distritbution of percent of mitochondrial reads per cell. ``` r library(scater) plotColData(sce.NeuroD1_filtered, x = "Sample", y = "subsets_Mito_percent") ``` <!-- --> --- ## Plotting QC data It also can be useful to capture the QC columns against each other as scatterplots ``` r library(scater) p1 <- plotColData(sce.NeuroD1_filtered, x = "sum", y = "detected") p2 <- plotColData(sce.NeuroD1_filtered, x = "sum", y = "subsets_Mito_percent") p3 <- plotColData(sce.NeuroD1_filtered, x = "detected", y = "subsets_Mito_percent") gridExtra::grid.arrange(p1, p2, p3, ncol = 3) ``` <!-- --> --- ## Removing low quality data At this point we may want to remove low quality data. Typically droplet filters on mitochondrial read content, total detect genes and total read counts are applied. ``` r qc.high_lib_size <- colData(sce.NeuroD1_filtered)$sum > 125000 qc.min_detected <- colData(sce.NeuroD1_filtered)$detected < 200 qc.mito <- colData(sce.NeuroD1_filtered)$subsets_Mito_percent > 25 discard <- qc.high_lib_size | qc.mito | qc.min_detected DataFrame(LibSize = sum(qc.high_lib_size), Detected = sum(qc.min_detected), MitoProp = sum(qc.mito), Total = sum(discard)) ``` ``` ## DataFrame with 1 row and 4 columns ## LibSize Detected MitoProp Total ## <integer> <integer> <integer> <integer> ## 1 13 473 1767 1792 ``` --- ## Removing low quality data We will add this back in the colData slot to perform some final QC plots before filtering ``` r colData(sce.NeuroD1_filtered) <- cbind(colData(sce.NeuroD1_filtered), DataFrame(toDiscard = discard)) p1 <- plotColData(sce.NeuroD1_filtered, x = "sum", y = "detected", colour_by = "toDiscard") p2 <- plotColData(sce.NeuroD1_filtered, x = "sum", y = "subsets_Mito_percent", colour_by = "toDiscard") p3 <- plotColData(sce.NeuroD1_filtered, x = "detected", y = "subsets_Mito_percent", colour_by = "toDiscard") gridExtra::grid.arrange(p1, p2, p3, ncol = 3) ``` 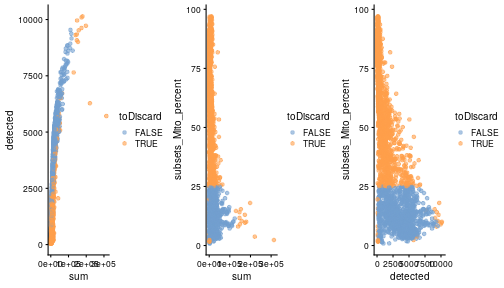<!-- --> --- ## Removing low quality data At this point we may want to remove the low quality cells from further analysis ``` r sce.NeuroD1_filtered_QCed <- sce.NeuroD1_filtered[, sce.NeuroD1_filtered$toDiscard %in% "FALSE"] sce.NeuroD1_filtered_QCed ``` ``` ## class: SingleCellExperiment ## dim: 39905 1676 ## metadata(1): Samples ## assays(1): counts ## rownames(39905): Gm26206 Xkr4 ... MT-TrnT MT-TrnP ## rowData names(3): ID Symbol Type ## colnames(1676): AAACCCACACAATGTC-1 AAACCCACACGAGAAC-1 ... ## TTTGGAGTCCGTAGGC-1 TTTGTTGGTCTAACTG-1 ## colData names(9): Sample Barcode ... total toDiscard ## reducedDimNames(0): ## mainExpName: NULL ## altExpNames(0): ``` --- # Log Normalisation As with our RNA-seq and genomics data we want to normalise for total library size and perfrom a log2 transformation. ``` r sce.NeuroD1_filtered_QCed <- logNormCounts(sce.NeuroD1_filtered_QCed) assayNames(sce.NeuroD1_filtered_QCed) ``` ``` ## [1] "counts" "logcounts" ``` --- ## Log Normalisation 2 An alternative approach for scRNA is to normalise accounting for potentially different cell-populations. As we dont know cell populations yet, a quick clustering of data followed by this normalisation using clusters is performed. ``` r library(scran) ``` ``` ## Warning: package 'scran' was built under R version 4.4.1 ``` ``` r clust.sce.NeuroD1_filtered_QCed <- quickCluster(sce.NeuroD1_filtered_QCed) sce.NeuroD1_filtered_trimmed <- computeSumFactors(sce.NeuroD1_filtered_QCed, cluster = clust.sce.NeuroD1_filtered_QCed) sce.NeuroD1_filtered_QCed <- logNormCounts(sce.NeuroD1_filtered_QCed) assayNames(sce.NeuroD1_filtered_QCed) ``` ``` ## [1] "counts" "logcounts" ``` --- ## Modeling the mean-variance relationship As with RNA-seq we want to account for mean-dispersion relationship. In Bioconductor we can use the *modelGeneVar* function to perform this for us. ``` r ## dec.NeuroD1_filtered_QCed <- modelGeneVar(sce.NeuroD1_filtered_QCed) ggplot(dec.NeuroD1_filtered_QCed, aes(x = mean, y = total)) + geom_point() ``` 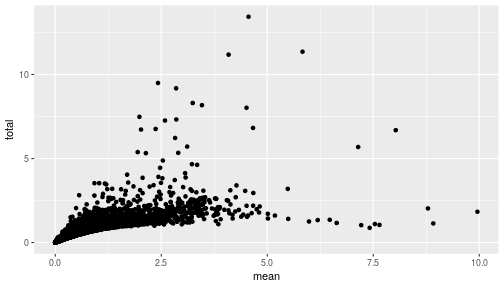<!-- --> --- ## Identifying top variable genes for PCA Having fit the mean-dispersion relationship we can extract the most variable genes for use in PCA dimension reduction. ``` r top.NeuroD1_filtered_QCed <- getTopHVGs(dec.NeuroD1_filtered_QCed, n = 3000) ``` --- ## PCA We can now perform a PCA on our data. The PCs will be used for clustering and 2D-representation of our data. ``` r set.seed(100) # See below. sce.NeuroD1_filtered_QCed <- fixedPCA(sce.NeuroD1_filtered_QCed, subset.row = top.NeuroD1_filtered_QCed) reducedDimNames(sce.NeuroD1_filtered_QCed) ``` ``` ## [1] "PCA" ``` --- ## PCA We can produce a quick plot of the PCs to see if there are any biases immediately evident in the PCs. ``` r plotReducedDim(sce.NeuroD1_filtered_QCed, dimred = "PCA", colour_by = "subsets_Mito_percent") ``` 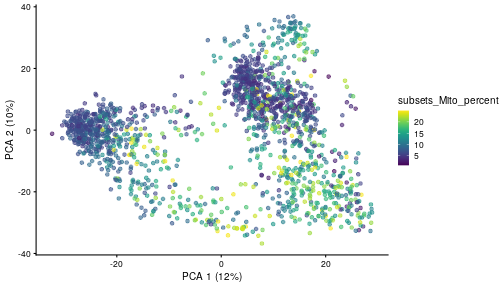<!-- --> --- ## Select PCs from PCA For further analysis we want to select the PCs which explain most of the data. To do this we can plot the variance explained by each PC and identify an elbow point. ``` r percent.var <- attr(reducedDim(sce.NeuroD1_filtered_QCed), "percentVar") plot(percent.var, log = "y", xlab = "PC", ylab = "Variance explained (%)") ``` 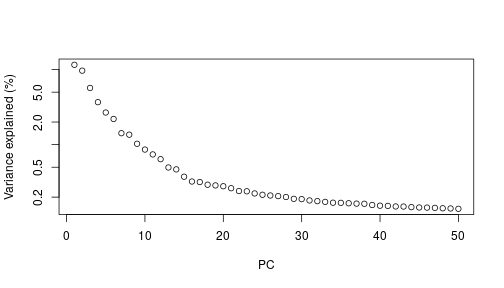<!-- --> ``` r # library(PCAtools) chosen.elbow <- findElbowPoint(percent.var) ``` --- ## Visualise in 2D with Tisne and UMAP With our PCs now calculated we can project these onto a 2D representation of the data. Here we will use two popular methods of TSNE and UMAP. ``` r sce.NeuroD1_filtered_QCed <- runTSNE(sce.NeuroD1_filtered_QCed, n_dimred = 30) sce.NeuroD1_filtered_QCed <- runUMAP(sce.NeuroD1_filtered_QCed, n_dimred = 30) reducedDimNames(sce.NeuroD1_filtered_QCed) ``` ``` ## [1] "PCA" "TSNE" "UMAP" ``` --- ## Visualise in 2D with Tisne and UMAP Witht the TSNE and UMAP we may first want to overlay some features of QC to see if this affects the data still. ``` r plotUMAP(sce.NeuroD1_filtered_QCed, colour_by = "subsets_Mito_percent") ``` <!-- --> ``` r plotTSNE(sce.NeuroD1_filtered_QCed, colour_by = "subsets_Mito_percent") ``` <!-- --> --- ## Clustering For clustering we can use the set of tools available in the [bluster](https://www.bioconductor.org/packages/release/bioc/html/bluster.html) package. Here we run the default graph based methods walktrap and louvain from the igraph package. ``` r require(bluster) ``` ``` ## Loading required package: bluster ``` ``` ## Warning: package 'bluster' was built under R version 4.4.1 ``` ``` r clust.louvain <- clusterCells(sce.NeuroD1_filtered_QCed, use.dimred = "PCA", BLUSPARAM = NNGraphParam(cluster.fun = "louvain", cluster.args = list(resolution = 0.8))) clust.default <- clusterCells(sce.NeuroD1_filtered_QCed, use.dimred = "PCA") ``` --- ## Clustering Once we have the clustering we will add them to the colData for the object and replot the UMAP ``` r colLabels(sce.NeuroD1_filtered_QCed) <- clust.louvain colData(sce.NeuroD1_filtered_QCed)$DefaultLabel <- clust.default plotUMAP(sce.NeuroD1_filtered_QCed, colour_by = "label") ``` <!-- --> --- ## Clustering We can now plot the mito percent against clusters to see if any clusters are driven by the mito percent of droplets. ``` r plotColData(sce.NeuroD1_filtered_QCed, x = "label", y = "subsets_Mito_percent", colour_by = "label") ``` <!-- --> --- ## Clustering We can then compare our overlaps between our clusterings in a simple heatmap. ``` r tab <- table(Walktrap = clust.default, Louvain = clust.louvain) rownames(tab) <- paste("Walktrap", rownames(tab)) colnames(tab) <- paste("louvain", colnames(tab)) library(pheatmap) pheatmap(log10(tab + 10), color = viridis::viridis(100), cluster_cols = FALSE, cluster_rows = FALSE) ``` 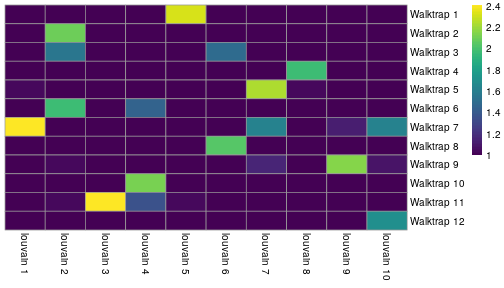<!-- --> --- ## Some ground truth The paper also provides their annotation of cells. We can read this in and overlay with our umap. ``` r eec_paper_meta <- read.delim("https://rubioinformatics.s3.amazonaws.com/scRNA_graduate/GSE224223_EEC_metadata.csv", sep = ";") nrd1_cells <- as.data.frame(eec_paper_meta[eec_paper_meta$Library == "Nd1", ]) nrd1_cells$X <- gsub("_2$", "", nrd1_cells$X) newMeta <- merge(colData(sce.NeuroD1_filtered_QCed), nrd1_cells, by.x = "Barcode", by.y = "X", all.x = TRUE, all.y = FALSE, order = FALSE) rownames(newMeta) <- newMeta$Barcode newMeta$All_Cell_Types[is.na(newMeta$All_Cell_Types)] <- "Filtered" colData(sce.NeuroD1_filtered_QCed) <- newMeta plotUMAP(sce.NeuroD1_filtered_QCed, colour_by = "All_Cell_Types") ``` 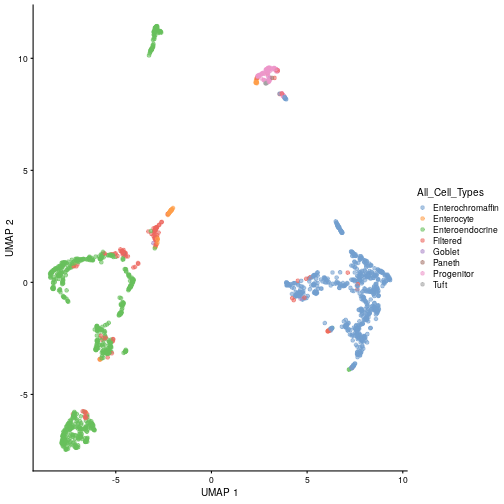<!-- --> --- ## Some ground truth We can also see how the annotation overlaps with our clusters. ``` r tab <- table(Walktrap = sce.NeuroD1_filtered_QCed$All_Cell_Types, Louvain = sce.NeuroD1_filtered_QCed$label) rownames(tab) <- paste("CellType", rownames(tab)) colnames(tab) <- paste("Leiden", colnames(tab)) library(pheatmap) pheatmap(log10(tab + 10), color = viridis::viridis(100), cluster_cols = FALSE, cluster_rows = FALSE) ``` 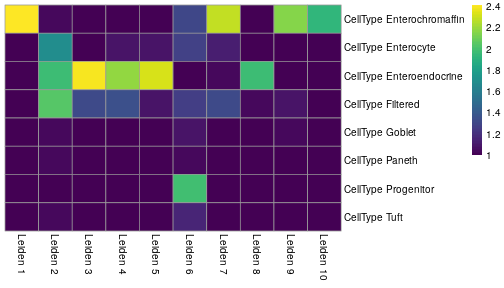<!-- --> --- ## Identify markers Now we have clusters defined we can identify genes which are differentially expressed within a cluster compared to other clusters. ``` r markers.sce.NeuroD1 <- scoreMarkers(sce.NeuroD1_filtered_QCed, sce.NeuroD1_filtered_QCed$label) chosen <- markers.sce.NeuroD1[["4"]] ordered <- chosen[order(chosen$mean.AUC, decreasing = TRUE), ] head(ordered[, 1:4]) # showing basic stats only, for brevity. ``` ``` ## DataFrame with 6 rows and 4 columns ## self.average other.average self.detected other.detected ## <numeric> <numeric> <numeric> <numeric> ## Sct 10.71205 7.178612 1.000000 0.994845 ## Scg2 5.92739 2.178339 1.000000 0.608673 ## Bace2 2.34958 0.500731 0.901961 0.280589 ## Etv1 2.57455 0.585889 0.941176 0.242338 ## Gpd2 2.30863 0.837072 0.960784 0.479837 ## Cyp4b1 2.55790 0.965280 0.934641 0.537153 ``` --- ## Plot markers We can then use the *plotExpression* function to visualise a gene's expression across clusters. ``` r plotExpression(sce.NeuroD1_filtered_QCed, features = head(rownames(ordered), n = 1), x = "label", colour_by = "label") ``` 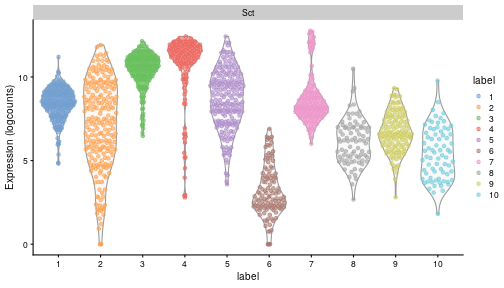<!-- --> ``` r saveRDS(sce.NeuroD1_filtered_QCed, file = "sce.NeuroD1_filtered_QCed.RDS") ``` --- class: inverse, center, middle # Seurat <html><div style='float:left'></div><hr color='#EB811B' size=1px width=720px></html> --- --- ## Working with CellRanger filtered matrix We can use Seurat to import our data as before for Bioconductor. ``` r library(Seurat) ``` ``` ## Loading required package: SeuratObject ``` ``` ## Loading required package: sp ``` ``` ## ## Attaching package: 'sp' ``` ``` ## The following object is masked from 'package:IRanges': ## ## %over% ``` ``` ## ## Attaching package: 'SeuratObject' ``` ``` ## The following object is masked from 'package:SummarizedExperiment': ## ## Assays ``` ``` ## The following object is masked from 'package:GenomicRanges': ## ## intersect ``` ``` ## The following object is masked from 'package:GenomeInfoDb': ## ## intersect ``` ``` ## The following object is masked from 'package:IRanges': ## ## intersect ``` ``` ## The following object is masked from 'package:S4Vectors': ## ## intersect ``` ``` ## The following object is masked from 'package:BiocGenerics': ## ## intersect ``` ``` ## The following objects are masked from 'package:base': ## ## intersect, t ``` ``` ## ## Attaching package: 'Seurat' ``` ``` ## The following object is masked from 'package:SummarizedExperiment': ## ## Assays ``` ``` r h5file <- "https://rubioinformatics.s3.amazonaws.com/scRNA_graduate/SRR_NeuroD1/outs/filtered_feature_bc_matrix.h5" local_h5file <- basename(h5file) download.file(h5file, local_h5file) Nd1T.mat <- Read10X_h5(filename = local_h5file) Nd1T.obj <- CreateSeuratObject(Nd1T.mat, project = "Nd1") ``` ``` ## Warning: Feature names cannot have underscores ('_'), replacing with dashes ## ('-') ``` --- ## The Seurat object The Seurat object contains now our information on Neurod1 droplets. ``` r Nd1T.obj ``` ``` ## An object of class Seurat ## 39905 features across 3468 samples within 1 assay ## Active assay: RNA (39905 features, 0 variable features) ## 1 layer present: counts ``` --- ## The Seurat object We can access rownames and column names in a similar manner as before. ``` r rownames(Nd1T.obj)[1:2] ``` ``` ## [1] "Gm26206" "Xkr4" ``` ``` r colnames(Nd1T.obj)[1:2] ``` ``` ## [1] "AAACCCACACAATGTC-1" "AAACCCACACCACTGG-1" ``` --- ## The Seurat object To access metadata we need to directly access the slot ``` r Nd1T.obj@meta.data[1:2, ] ``` ``` ## orig.ident nCount_RNA nFeature_RNA ## AAACCCACACAATGTC-1 Nd1 23320 4776 ## AAACCCACACCACTGG-1 Nd1 1944 176 ``` --- ## The Seurat object We can also review the active assay and active cell-barcode identifiers. ``` r Nd1T.obj@active.assay ``` ``` ## [1] "RNA" ``` ``` r Nd1T.obj@active.ident[1:2] ``` ``` ## AAACCCACACAATGTC-1 AAACCCACACCACTGG-1 ## Nd1 Nd1 ## Levels: Nd1 ``` --- ## The Seurat object The get access to the assays we can pull directly from the relevent slots. ``` r Nd1T.obj@assays$RNA$counts[1:2, 1:2] ``` ``` ## 2 x 2 sparse Matrix of class "dgCMatrix" ## AAACCCACACAATGTC-1 AAACCCACACCACTGG-1 ## Gm26206 . . ## Xkr4 . . ``` --- ## Gather QC As with Biocondcutor we must again identify mitochondrial genes. ``` r mito.genes <- grep("^MT", rownames(Nd1T.obj), value = T) mito.genes[1:10] ``` ``` ## [1] "MT-TrnF" "MT-mt-Rnr1" "MT-TrnV" "MT-mt-Rnr2" "MT-TrnL1" ## [6] "MT-ND1" "MT-TrnI" "MT-TrnQ" "MT-TrnM" "MT-ND2" ``` --- ## Gather QC We can the use *PercentageFeatureSet* function to identify percent of mito reads in each cell and the *AddMetaData* function to add this to the Seurat object. ``` r percent.mt <- PercentageFeatureSet(Nd1T.obj, features = mito.genes) Nd1T.obj <- AddMetaData(Nd1T.obj, metadata = percent.mt, col.name = "percent.mt") head(Nd1T.obj@meta.data) ``` ``` ## orig.ident nCount_RNA nFeature_RNA percent.mt ## AAACCCACACAATGTC-1 Nd1 23320 4776 14.399657 ## AAACCCACACCACTGG-1 Nd1 1944 176 81.121399 ## AAACCCACACGAGAAC-1 Nd1 76762 7564 9.710534 ## AAACCCAGTGTCCGGT-1 Nd1 17440 4177 30.928899 ## AAACGAAAGCCTCGTG-1 Nd1 9512 2187 29.447014 ## AAACGAAAGCCTTGAT-1 Nd1 11320 3181 14.187279 ``` --- ## Plot QC We can then review QC metrics across our droplets. ``` r VlnPlot(Nd1T.obj, features = c("nFeature_RNA", "nCount_RNA", "percent.mt"), ncol = 3) ``` ``` ## Warning: Default search for "data" layer in "RNA" assay yielded no results; ## utilizing "counts" layer instead. ``` <!-- --> --- ## Plot QC As well as plot QC metrics against each other. ``` r plot1 <- FeatureScatter(Nd1T.obj, feature1 = "nCount_RNA", feature2 = "percent.mt") plot2 <- FeatureScatter(Nd1T.obj, feature1 = "nCount_RNA", feature2 = "nFeature_RNA") plot3 <- FeatureScatter(Nd1T.obj, feature1 = "nFeature_RNA", feature2 = "percent.mt") plot1 + plot2 + plot3 ``` <!-- --> --- ## Subset by QC We use the subset function to remove low quality cells from our object. ``` r Nd1T.obj.filt <- subset(Nd1T.obj, subset = nCount_RNA < 125000 & percent.mt < 25 & nFeature_RNA > 200) Nd1T.obj.filt ``` ``` ## An object of class Seurat ## 39905 features across 1676 samples within 1 assay ## Active assay: RNA (39905 features, 0 variable features) ## 1 layer present: counts ``` --- ## Normalise data We will now normalise our data by scaling to library size and applying a log2 transformation. ``` r Nd1T.obj.filt <- NormalizeData(Nd1T.obj.filt) ``` ``` ## Normalizing layer: counts ``` --- ## Variable features As with Bioc we will now identify variable features. Here the mean-variance relationship is accounted for by applying a VST transformation. ``` r Nd1T.obj.filt <- FindVariableFeatures(Nd1T.obj.filt, selection.method = "vst", nfeatures = 3000) ``` ``` ## Finding variable features for layer counts ``` ``` r top10 <- head(VariableFeatures(Nd1T.obj.filt), 10) top10 ``` ``` ## [1] "Olfm4" "Iapp" "H2ac23" "Dmbt1" "Ube2c" "Asic5" "Ifitm3" "Reg3b" ## [9] "Hmgb2" "Reg3g" ``` --- ## Variable features We can then plot these features using the **VariableFeaturePlot** function. ``` r plot1 <- VariableFeaturePlot(Nd1T.obj.filt) plot2 <- LabelPoints(plot = plot1, points = top10, repel = TRUE) ``` ``` ## When using repel, set xnudge and ynudge to 0 for optimal results ``` ``` r plot2 ``` ``` ## Warning in scale_x_log10(): log-10 transformation introduced infinite values. ``` <!-- --> --- ## Scaling data In Bioconductor the scaling of data was done inside the PCA functions. Here we apply the scaling using the **ScaleData** function. ``` r all.genes <- rownames(Nd1T.obj.filt) Nd1T.obj.filt <- ScaleData(Nd1T.obj.filt, features = all.genes) ``` ``` ## Centering and scaling data matrix ``` ``` r Nd1T.obj.filt@assays ``` ``` ## $RNA ## Assay (v5) data with 39905 features for 1676 cells ## Top 10 variable features: ## Olfm4, Iapp, H2ac23, Dmbt1, Ube2c, Asic5, Ifitm3, Reg3b, Hmgb2, Reg3g ## Layers: ## counts, data, scale.data ``` --- ## PCA Now we have our scaled data we can apply PCA and identify how many PCs to use in downstream analysis. ``` r Nd1T.obj.filt <- RunPCA(Nd1T.obj.filt, features = VariableFeatures(object = Nd1T.obj)) ``` ``` ## PC_ 1 ## Positive: Mir6236, Apoa1, Bnip5, Trpm2, Apoa4, Apob, Ada, Nts, Cyp3a11, Gm53845 ## Vgf, Klf4, Col27a1, Gm57596, Rfx6, Prss30, Sst, Pmp22, Gm32584, Rasal2 ## Ly6m, Nrp1, Gm50599, Slc38a11, Adgrd1, Ahnak, Slc26a4, Afp, Slc28a2, Apol7a ## Negative: Rps2, Rps8, Rpl12, Rpl27a, Eef1b2, Rplp0, Gvin-ps1, Rpl32, Rps15a, Rpl22 ## Cps1, Rpl18a, Rpl13, Rps20, Rps16, Rpl35, Rps7, Rps28, Rpl23, Rps3 ## Rpl28, Rps23, Rps4x, Rpl35a, Rps12, Rps24, Rps10, Rps27a, Nme2, Rpl30 ## PC_ 2 ## Positive: Rbp4, Scgn, Gm11992, Ptma, Bambi, Etv1, Parm1, Cltrn, Isl1, Qpct ## Tmem176a, Kcnk16, Crp, Gm53467, Pzp, Gnai1, Rps3a1, Galnt13, Dner, Rpl6 ## Rps24, Rfxap, Cpn1, Tril, Cyp2j5, Smbd1, Rpl12, Kcnj6, Agr2, Tff3 ## Negative: Spink1, Clec2h, Mep1b, Ace2, Guca2b, Slc51a, Apoa1, Ces2e, Ggt1, Lgals3 ## Alpi, Apoa4, Clca4b, Ifi27l2b, Dpep1, Mogat2, Adh6a, H2-T3, Ace, Treh ## Maf, Xdh, Cndp2, Maoa, Apol10a, Fabp2, H2-T26, Mall, Arg2, Dnase1 ## PC_ 3 ## Positive: Rbp4, Scgn, Isl1, Gm11992, Gsto1, Cyp2j6, Etv1, Bambi, Gpd2, Ugt2b34 ## S100a11, Abcc8, St3gal4, Cltrn, Fabp2, Parm1, Ghrl, Gnai1, Kcnk16, Pzp ## Galnt13, Crp, Higd1a, Kcnj6, Ano6, Acvr1c, Plcxd3, Tril, Slc41a2, Cds1 ## Negative: Tpbg, Slc38a11, Lmx1a, Rasal2, Ccnd2, Krt19, Afp, Trpa1, Shisa2, Pla2g7 ## Amigo2, Trpm2, Serpinf2, Lypd8l, Sorl1, Slc5a4b, Gm53845, Cwh43, Rpp25, Grik4 ## Adh1, 9330154J02Rik, Rab3c, Lonrf1, Fabp3, Apoa5, Nars2, Slc5a9, Ptgr1, Tmem238l ## PC_ 4 ## Positive: Top2a, Nusap1, Gvin-ps1, Clca3b, Aurkb, Cenpf, D17H6S56E-5, Ckap2l, Hmmr, Mir6236 ## H1f5, Cenpe, Hmgb2, Cdca2, Melk, Esco2, Cd200l2, Stmn1, Bub1b, Prc1 ## Uhrf1, Ckap2, Ncapg, H2ac23, Cdca7, Cdca3, Slfn9, Cenpw, Ccnb2, Cenpm ## Negative: Tff3, Agr3, Ttr, Tmem176a, Casp6, Agr2, Uqcr11, Uqcrb, Pzp, Ppia ## Ndufa4, Glod5, D930028M14Rik, Homer2, Tle1, Cyp2j6, Cpn1, Uqcrq, Onecut3, Trf ## Atp5me, Crp, Qpct, Ndufb10, Lypd8, Cox5b, Tmed6, Cyb5a, Rpl19, Atp5mj ## PC_ 5 ## Positive: Agr2, Agr3, Mgll, Phgr1, Onecut3, Serpina1e, Tmsb4x, Pzp, Homer2, Klf4 ## Tle1, Hspb1, Galnt13, D930028M14Rik, Cit, Crp, S100a10, Cpn1, Gm12511, Ckb ## Serpina1b, Kcnk16, Lamb3, Gm53467, Slc43a2, Slc41a2, Glod5, Cyp2j5, Fgl2, Trpm2 ## Negative: Mrln, Scarb1, Rgs4, Phlda1, Rfx6, Rbp2, 2010204K13Rik, Fam167a, Ass1, 4933407L21Rik ## Nherf4, Trpc5, Slfn10-ps, Isl1, Plcxd3, Gamt, Th, Sfrp5, Prkg2, Fmo1 ## Gm32255, Plxnc1, Cth, Pls3, Ntrk1, Gsto1, Gm54263, Acvr1c, Alpl, Prps1 ``` ``` r ElbowPlot(Nd1T.obj.filt, ndims = 50) ``` <!-- --> --- ## UMAP We can then create a 2D representation of our data using a UMAP . ``` r Nd1T.obj.filt <- RunUMAP(Nd1T.obj.filt, dims = 1:30) ``` ``` ## Warning: The default method for RunUMAP has changed from calling Python UMAP via reticulate to the R-native UWOT using the cosine metric ## To use Python UMAP via reticulate, set umap.method to 'umap-learn' and metric to 'correlation' ## This message will be shown once per session ``` ``` ## 14:11:36 UMAP embedding parameters a = 0.9922 b = 1.112 ``` ``` ## 14:11:36 Read 1676 rows and found 30 numeric columns ``` ``` ## 14:11:36 Using Annoy for neighbor search, n_neighbors = 30 ``` ``` ## 14:11:36 Building Annoy index with metric = cosine, n_trees = 50 ``` ``` ## 0% 10 20 30 40 50 60 70 80 90 100% ``` ``` ## [----|----|----|----|----|----|----|----|----|----| ``` ``` ## **************************************************| ## 14:11:36 Writing NN index file to temp file /tmp/RtmpQhrJb1/file17935da4290f ## 14:11:36 Searching Annoy index using 1 thread, search_k = 3000 ## 14:11:36 Annoy recall = 100% ## 14:11:37 Commencing smooth kNN distance calibration using 1 thread with target n_neighbors = 30 ## 14:11:39 Initializing from normalized Laplacian + noise (using RSpectra) ## 14:11:39 Commencing optimization for 500 epochs, with 65880 positive edges ## 14:11:41 Optimization finished ``` --- ## UMAP And we plot this using the *FeaturePlot* function overlaying the percent.mt. ``` r FeaturePlot(Nd1T.obj.filt, reduction = "umap", features = "percent.mt") ``` <!-- --> --- ## Clustering We can then perform a louvain clustering using the *FindNeighbours* and *FindClusters* functions. The cluster labels will be stored in active.idents slots. ``` r Nd1T.obj.filt <- FindNeighbors(Nd1T.obj.filt, dims = 1:30) ``` ``` ## Computing nearest neighbor graph ``` ``` ## Computing SNN ``` ``` r Nd1T.obj.filt <- FindClusters(Nd1T.obj.filt, resolution = 0.7) ``` ``` ## Modularity Optimizer version 1.3.0 by Ludo Waltman and Nees Jan van Eck ## ## Number of nodes: 1676 ## Number of edges: 54522 ## ## Running Louvain algorithm... ## Maximum modularity in 10 random starts: 0.8620 ## Number of communities: 12 ## Elapsed time: 0 seconds ``` ``` r Nd1T.obj.filt@active.ident ``` ``` ## AAACCCACACAATGTC-1 AAACCCACACGAGAAC-1 AAACGAAAGCCTTGAT-1 AAACGAACATGTCAGT-1 ## 6 5 1 1 ## AAACGAAGTAGACGTG-1 AAACGAAGTAGCTGTT-1 AAACGAAGTGTATACC-1 AAACGCTAGAGTTGAT-1 ## 6 6 4 2 ## AAACGCTCATCGCTCT-1 AAAGAACCAGACTCTA-1 AAAGAACCATATCGGT-1 AAAGAACCATCCTCAC-1 ## 1 5 1 6 ## AAAGAACGTACCTAAC-1 AAAGAACGTTGGACCC-1 AAAGAACTCGTGCACG-1 AAAGGATCATGTTCGA-1 ## 4 2 5 2 ## AAAGGTACAACTTGGT-1 AAAGGTACATAGGCGA-1 AAAGTCCCATGGCACC-1 AAAGTCCTCTTCGATT-1 ## 2 0 5 0 ## AAAGTGAAGATGACAT-1 AAATGGAAGTGTAGTA-1 AAATGGACATGCTGCG-1 AACAAAGGTCCGGTGT-1 ## 2 9 3 6 ## AACAAAGTCGCAATTG-1 AACAAGAGTGAGTGAC-1 AACAGGGCATGTCAGT-1 AACCAACAGCCTGTGC-1 ## 0 2 8 2 ## AACCAACAGGTCATAA-1 AACCAACGTTGCGGAA-1 AACCAACTCCTATGGA-1 AACCACAAGGTGGTTG-1 ## 0 1 5 3 ## AACCACAAGTAACCTC-1 AACCACACATCCAATG-1 AACCACAGTCCAGGTC-1 AACCACAGTCGACGCT-1 ## 6 5 0 1 ## AACCCAAAGATGAACT-1 AACCCAATCCAGCAAT-1 AACCTGACACTCTCGT-1 AACCTGAGTGCATGAG-1 ## 8 3 9 2 ## AACGAAAAGGGACTGT-1 AACGAAACAAGAAATC-1 AACGGGACACTCACTC-1 AACGGGATCTCGCTCA-1 ## 0 0 0 9 ## AACGTCAAGCGATGGT-1 AACGTCAGTGTCCACG-1 AACGTCATCTGGTCAA-1 AACTTCTTCATCACTT-1 ## 5 6 8 9 ## AAGAACAAGGGCAGAG-1 AAGACTCAGAAATTGC-1 AAGACTCAGCAGCGAT-1 AAGACTCCAACCGTGC-1 ## 3 1 4 10 ## AAGACTCCAAGTGCAG-1 AAGACTCTCCCGTAAA-1 AAGACTCTCTAAGGAA-1 AAGATAGAGGGCAATC-1 ## 1 9 4 0 ## AAGCATCCACATGTTG-1 AAGCATCTCAGATTGC-1 AAGCATCTCTCGCAGG-1 AAGCGTTAGGCCGCTT-1 ## 2 0 2 3 ## AAGCGTTAGGGCAGTT-1 AAGCGTTCATTCTTCA-1 AAGGAATAGCCTAGGA-1 AAGGTAAGTAATCAAG-1 ## 2 3 0 1 ## AAGTCGTAGGTTCAGG-1 AAGTCGTTCATCCCGT-1 AAGTCGTTCTGTTGGA-1 AAGTGAAAGCCGGATA-1 ## 7 0 5 3 ## AAGTGAACACCCTGAG-1 AAGTGAAGTTTGGCTA-1 AATCACGAGAGCGACT-1 AATCACGCAAGCGCTC-1 ## 4 8 4 11 ## AATCACGCAATGAGCG-1 AATCACGCAGACAAGC-1 AATCGACGTGCGGATA-1 AATCGACTCATCGTAG-1 ## 0 4 0 3 ## AATCGACTCTAAGGAA-1 AATGACCAGGGTGAAA-1 AATGACCCAGAGTAAT-1 AATGACCGTCTGTAGT-1 ## 4 1 2 8 ## AATGACCGTGGTCCCA-1 AATGCCACATAACTCG-1 AATGCCATCCGGCTTT-1 AATGGAAAGCGTATGG-1 ## 9 1 7 1 ## AATGGAAGTCAGTTTG-1 AATTCCTTCTCCCATG-1 AATTTCCGTCATATGC-1 ACAAAGAAGCTACTGT-1 ## 0 3 3 2 ## ACAAAGACAACTCATG-1 ACAAAGACAGATAAAC-1 ACAAAGAGTACTGCGC-1 ACAACCACAACACTAC-1 ## 4 5 6 6 ## ACAACCACAATTCGTG-1 ACAACCACACCCAACG-1 ACAACCATCAAGCCAT-1 ACAACCATCCTACAAG-1 ## 4 7 11 4 ## ACAAGCTAGCCAAGCA-1 ACAAGCTCACAGAGAC-1 ACAAGCTCACCTGATA-1 ACAAGCTCAGCGTACC-1 ## 0 9 0 0 ## ACAAGCTGTTATTCCT-1 ACAAGCTTCGGTTAGT-1 ACACAGTAGCACGTCC-1 ACACCAATCATCCTGC-1 ## 2 6 8 1 ## ACACCAATCCACCTGT-1 ACACGCGCAATAGGGC-1 ACACTGAGTGCCAAGA-1 ACAGAAACACCGAATT-1 ## 5 0 0 5 ## ACAGAAACAGAGGACT-1 ACAGCCGAGGCCGCTT-1 ACAGGGAAGAAACTGT-1 ACAGGGAGTTGAATCC-1 ## 1 4 10 9 ## ACATCCCAGCAAGCCA-1 ACATCCCAGGCTAGCA-1 ACATCCCCATCAGTGT-1 ACATGCACATCTTTCA-1 ## 0 1 6 4 ## ACATGCAGTACTGCGC-1 ACATGCAGTCCTGGGT-1 ACATGCAGTGCATACT-1 ACATTTCAGTCACTGT-1 ## 5 4 1 5 ## ACATTTCGTATTTCTC-1 ACATTTCTCCGCACTT-1 ACATTTCTCGTGTGGC-1 ACCAAACAGACCTCAT-1 ## 5 2 2 1 ## ACCAACAAGCCGCTTG-1 ACCAACAGTCGGCTAC-1 ACCACAAGTCGAAGCA-1 ACCACAAGTTTGGAAA-1 ## 6 0 8 2 ## ACCACAATCACAAGGG-1 ACCATTTAGACGACTG-1 ACCATTTAGGTACAGC-1 ACCATTTAGTCGTCTA-1 ## 7 1 11 2 ## ACCATTTGTAGTCCTA-1 ACCATTTGTATGTCCA-1 ACCATTTGTGACTGTT-1 ACCCTCATCCTCGCAT-1 ## 7 1 0 2 ## ACCTACCCAGCGTTGC-1 ACCTACCTCAAGCTTG-1 ACCTACCTCGACGACC-1 ACCTACCTCGTTTACT-1 ## 3 6 3 6 ## ACCTGAAAGCAACAGC-1 ACCTGAACAATTGCGT-1 ACCTGAAGTCTGGTTA-1 ACGATCAGTCCGTTTC-1 ## 5 2 2 10 ## ACGATGTCAAGTTCGT-1 ACGATGTCATGTGCTA-1 ACGATGTTCGAAATCC-1 ACGATGTTCTATCGCC-1 ## 0 2 0 5 ## ACGCACGCAGGTTACT-1 ACGGAAGCAAGGCCTC-1 ACGGAAGCAGGTATGG-1 ACGGGTCAGGCTGGAT-1 ## 6 1 7 3 ## ACGGGTCAGTAGCTCT-1 ACGGGTCGTCACTCAA-1 ACGGTCGAGCGCTGAA-1 ACGGTCGGTATCGATC-1 ## 7 0 3 9 ## ACGGTTAAGCGTGCCT-1 ACGGTTAAGCTCGAAG-1 ACGGTTAGTCGTAATC-1 ACGTACAAGCAGGTCA-1 ## 10 4 4 0 ## ACGTACATCGTTGTGA-1 ACGTAGTAGGGCGAGA-1 ACGTAGTCATCACAGT-1 ACGTAGTGTCCTATAG-1 ## 2 6 0 0 ## ACGTCCTAGCATCAGG-1 ACGTCCTGTGGCTACC-1 ACGTCCTGTTAAGGGC-1 ACGTTCCCACTATGTG-1 ## 1 4 1 5 ## ACTATCTGTGGAGAAA-1 ACTATGGGTTCTCGCT-1 ACTATTCTCAAACCCA-1 ACTCCCATCTAGCATG-1 ## 3 0 1 2 ## ACTCTCGGTCGTGGTC-1 ACTCTCGGTGGAGAAA-1 ACTGATGCAAATGGAT-1 ACTGCAAAGGACTGGT-1 ## 7 2 5 3 ## ACTGCAAAGGTATCTC-1 ACTGTCCCATCTGGGC-1 ACTGTCCGTCGCACGT-1 ACTGTGACACCCTAAA-1 ## 3 8 0 1 ## ACTGTGATCTAACACG-1 ACTTAGGAGCAGCGAT-1 ACTTAGGAGTGAGTTA-1 ACTTAGGCAAGTGGTG-1 ## 2 6 2 1 ## ACTTAGGGTCCAGCCA-1 ACTTATCGTATCGCTA-1 ACTTATCGTTAGAAGT-1 ACTTATCTCCTGTAGA-1 ## 3 5 0 1 ## ACTTCCGGTACTGCGC-1 ACTTCCGTCTCCCATG-1 ACTTCGCCAGGTTCAT-1 ACTTCGCGTAGAGACC-1 ## 3 3 0 4 ## ACTTTGTAGACGCTCC-1 ACTTTGTAGTAAATGC-1 ACTTTGTGTAGAATGT-1 AGAAATGGTCATCGGC-1 ## 2 1 1 3 ## AGAAATGGTTCAGGTT-1 AGAACAAAGAGTGTGC-1 AGAACAAAGGGCAATC-1 AGAACCTGTAGACGGT-1 ## 9 8 1 4 ## AGAACCTGTGCCTTTC-1 AGAACCTTCAGTCTTT-1 AGAAGTAAGAAGCGCT-1 AGAAGTACATGGGCAA-1 ## 3 3 8 6 ## AGACAAACATCGCCTT-1 AGACAAATCCCGAATA-1 AGACACTGTAACCCTA-1 AGACACTTCCTACCAC-1 ## 5 2 1 5 ## AGACAGGCATCGTGCG-1 AGACCATAGCGACCCT-1 AGACCCGTCGAACACT-1 AGACCCGTCGTTGTTT-1 ## 4 0 1 0 ## AGAGAATCAACTGGTT-1 AGAGAATTCCCGTGTT-1 AGAGAGCTCAGATGCT-1 AGAGCAGAGGTTCTTG-1 ## 1 2 4 0 ## AGAGCAGAGTTTGGCT-1 AGAGCAGCAAGGACAC-1 AGAGCAGCAGCACCCA-1 AGAGCAGGTGATGTAA-1 ## 1 4 11 8 ## AGAGCCCAGCCACAAG-1 AGATAGAAGGATGTTA-1 AGATCCAAGCCTCGTG-1 AGATCCATCAGCTTCC-1 ## 0 0 0 1 ## AGATCGTCATTGAGCT-1 AGATCGTGTAGCGAGT-1 AGATGAATCAAACGAA-1 AGATGCTAGCATCAAA-1 ## 1 4 1 1 ## AGATGCTAGGTGCAGT-1 AGATGCTAGTAGAGTT-1 AGATGCTCATGTCAGT-1 AGATGCTGTACTCGCG-1 ## 1 6 5 2 ## AGCATCAGTCCTGTTC-1 AGCCAATGTCATTCCC-1 AGCCAGCGTTAGGCTT-1 AGCGCCAAGAGGGCGA-1 ## 2 6 4 5 ## AGCGCCACATAATCGC-1 AGCGCCAGTATAGCTC-1 AGCGTATCAAATTGGA-1 AGCGTATGTTGCCATA-1 ## 1 5 2 6 ## AGCGTATTCTGGCCGA-1 AGCGTCGCATATCGGT-1 AGCTACAGTCGGATTT-1 AGCTACATCGTCAAAC-1 ## 10 0 0 0 ## AGCTCAAAGCAGATAT-1 AGCTCAAAGTCTGTAC-1 AGCTCAACACCTTCGT-1 AGCTCAATCGTTCGCT-1 ## 1 7 10 6 ## AGCTTCCAGAAGCTCG-1 AGCTTCCAGGTGCCTC-1 AGCTTCCTCAAGATAG-1 AGCTTCCTCGTGTTCC-1 ## 4 1 2 10 ## AGGAAATAGAAGCTGC-1 AGGAAATTCATTCCTA-1 AGGAATATCTGTAACG-1 AGGACGACATGGGAAC-1 ## 8 6 0 3 ## AGGACTTAGTCCTACA-1 AGGACTTCAGAGGACT-1 AGGAGGTAGGAGCAAA-1 AGGAGGTGTTATGGTC-1 ## 0 4 7 0 ## AGGATAAGTGTAGCAG-1 AGGATCTAGACGTCCC-1 AGGATCTCAACGATCT-1 AGGCCACAGTTCATCG-1 ## 4 1 11 8 ## AGGCCACCATCAGTCA-1 AGGCCACCATGGGTCC-1 AGGGCCTAGTACAACA-1 AGGGCCTAGTTGCATC-1 ## 6 0 6 3 ## AGGGCCTCAGATTAAG-1 AGGGCCTTCCGTTGAA-1 AGGGCCTTCGTAGGGA-1 AGGGCTCAGGACCCAA-1 ## 8 2 0 1 ## AGGGCTCCAGCTCCTT-1 AGGGCTCGTAGCGCCT-1 AGGGCTCTCTCGACGG-1 AGGGTCCTCGTTACCC-1 ## 0 6 0 10 ## AGGGTGAAGCATACTC-1 AGGTAGGCACGTAACT-1 AGGTAGGGTACGTACT-1 AGGTAGGGTGAGCCAA-1 ## 0 1 1 7 ## AGGTCTATCGATTTCT-1 AGGTTACGTACAGGTG-1 AGTAACCCAGACTGCC-1 AGTACCAGTCGCGTTG-1 ## 5 0 2 1 ## AGTACTGAGGGTTTCT-1 AGTACTGCACTTGACA-1 AGTACTGGTATGGTAA-1 AGTAGTCGTGAGTGAC-1 ## 0 8 9 9 ## AGTAGTCGTTGTGTTG-1 AGTCAACAGAGTGTGC-1 AGTCAACCACTAACCA-1 AGTCACAAGCGATGAC-1 ## 3 5 4 10 ## AGTCATGCAGCACAAG-1 AGTCATGTCTCCCAAC-1 AGTCTCCCATGCCGAC-1 AGTCTCCCATGGACAG-1 ## 3 3 5 9 ## AGTGATCAGGACGCTA-1 AGTTAGCGTTAGGACG-1 AGTTCCCGTTACCGTA-1 ATACCGATCCGTAGTA-1 ## 3 4 1 5 ## ATACCTTGTCGATTCA-1 ATACCTTTCTAGAGCT-1 ATACCTTTCTCCCATG-1 ATAGACCCACTCGATA-1 ## 0 0 3 4 ## ATAGACCGTCATCGCG-1 ATAGACCTCATGCCAA-1 ATAGAGACAAGAATGT-1 ATAGGCTGTAACGCGA-1 ## 0 0 0 4 ## ATAGGCTTCTAGCCAA-1 ATATCCTCAGGCGATA-1 ATCACAGCACCGTGCA-1 ATCACAGCACGATAGG-1 ## 1 0 3 11 ## ATCACGAAGGTACAAT-1 ATCACGATCGCTAGCG-1 ATCACTTAGGGTTTCT-1 ATCACTTTCCTAAGTG-1 ## 5 0 0 1 ## ATCAGGTAGGGTGGGA-1 ATCATTCAGCAGGTCA-1 ATCATTCCATAGACTC-1 ATCATTCGTGACTAAA-1 ## 10 2 3 0 ## ATCATTCGTTCGTTCC-1 ATCCACCAGAGTTCGG-1 ATCCACCCAGTGGCTC-1 ATCCACCGTCATCCCT-1 ## 7 4 5 3 ## ATCCACCGTGTGACCC-1 ATCCATTCAAGCGATG-1 ATCCATTGTGCCCGTA-1 ATCCATTTCGGCATAT-1 ## 3 11 4 2 ## ATCCCTGAGGTAAGGA-1 ATCCCTGCAGCACCCA-1 ATCCCTGCATGTGACT-1 ATCCCTGTCTGAGATC-1 ## 6 6 1 5 ## ATCCGTCCAAGCTGTT-1 ATCCGTCGTCTGTAGT-1 ATCCGTCGTCTTGGTA-1 ATCCGTCTCTGAATGC-1 ## 7 9 3 3 ## ATCCTATAGTCGGGAT-1 ATCCTATCAAGTGATA-1 ATCCTATTCGTGCATA-1 ATCGATGAGTTAACAG-1 ## 0 2 1 0 ## ATCGATGCACCAGACC-1 ATCGATGGTGGGAGAG-1 ATCGCCTCAATGTCAC-1 ATCGGATGTAGTATAG-1 ## 0 1 0 0 ## ATCGTCCCAAGACCTT-1 ATCGTCCCACCGTACG-1 ATCTCTAGTTTCGTAG-1 ATCTTCACACAATCTG-1 ## 2 2 3 3 ## ATCTTCACACGCCACA-1 ATCTTCACAGACCCGT-1 ATGAAAGCATTGCTTT-1 ATGAAAGTCAAAGAAC-1 ## 4 0 0 3 ## ATGAAAGTCAGGGTAG-1 ATGAAAGTCATCTACT-1 ATGAAAGTCTCTTAAC-1 ATGACCAAGCGCGTTC-1 ## 2 0 0 2 ## ATGACCAAGGAACTCG-1 ATGACCACAAGTGCAG-1 ATGACCACATTCACAG-1 ATGACCAGTCGTTGCG-1 ## 5 0 7 2 ## ATGACCATCCCGAACG-1 ATGAGGGGTACCTAAC-1 ATGAGTCCAAAGACGC-1 ATGAGTCTCTGTACAG-1 ## 6 6 3 0 ## ATGATCGTCAAATGAG-1 ATGCCTCAGGTGCTGA-1 ATGCCTCTCACGGGAA-1 ATGCGATTCGTTCTCG-1 ## 1 8 0 11 ## ATGGATCGTGTAGGAC-1 ATGGATCTCCATCCGT-1 ATGGGAGGTGGTGATG-1 ATGGGTTAGCAACCAG-1 ## 0 1 3 0 ## ATGGTTGGTCAACACT-1 ATGGTTGGTTATCTGG-1 ATGGTTGTCAAGCTGT-1 ATGGTTGTCCCGATCT-1 ## 1 5 5 2 ## ATGGTTGTCCGCTAGG-1 ATGTCTTCACGGTCTG-1 ATGTCTTCACTGATTG-1 ATGTCTTCATCGCTAA-1 ## 2 4 6 9 ## ATGTCTTGTCATTCCC-1 ATTACTCTCCGGCTTT-1 ATTATCCAGACATAGT-1 ATTATCCGTATCGATC-1 ## 7 0 0 0 ## ATTATCCTCCCAAGTA-1 ATTCATCAGGGTAGCT-1 ATTCCATTCCGTCCTA-1 ATTCCTATCTAGGCCG-1 ## 0 3 2 0 ## ATTCGTTCAGAGTAAT-1 ATTCGTTTCGAGAATA-1 ATTCTACAGCTCGAAG-1 ATTCTACTCACCGGGT-1 ## 1 0 5 1 ## ATTCTTGAGGCAGGGA-1 ATTCTTGTCAACCTCC-1 ATTGGGTTCGTAGGGA-1 ATTGTTCCATCGAAGG-1 ## 8 6 1 9 ## ATTGTTCGTCCACGCA-1 ATTGTTCTCACCGACG-1 ATTGTTCTCTCCATAT-1 ATTTACCAGGGCGAGA-1 ## 5 8 3 8 ## ATTTACCAGTACTGTC-1 ATTTACCGTTGCATTG-1 ATTTCACAGTCCCAGC-1 ATTTCACGTTGGTAGG-1 ## 2 0 1 5 ## ATTTCTGAGAGCACTG-1 CAAAGAAGTGCCTACG-1 CAAAGAATCGTAGTGT-1 CAACAACAGTAGAGTT-1 ## 8 8 5 5 ## CAACAACTCTTACCAT-1 CAACAGTTCTCCCATG-1 CAACCAAAGATCCAAA-1 CAACCAAAGTGCACTT-1 ## 8 3 6 7 ## CAACCTCAGTGCAACG-1 CAACCTCGTATTTCGG-1 CAACCTCGTTGTCAGT-1 CAACGATAGATTGTGA-1 ## 7 11 4 1 ## CAACGATCAACGTATC-1 CAACGATTCCACTGGG-1 CAACGATTCCCGTGAG-1 CAACGATTCCGAAGGA-1 ## 8 5 1 2 ## CAACGGCGTAACACCT-1 CAAGACTCACTGAGTT-1 CAAGAGGAGCTACAAA-1 CAAGAGGCAACACGAG-1 ## 3 0 5 0 ## CAAGAGGGTGGGACAT-1 CAAGCTAAGACGGAAA-1 CAAGCTAGTCTCCTGT-1 CAAGCTAGTGCATGTT-1 ## 4 1 8 2 ## CAAGCTATCCTTCGAC-1 CAAGGGAAGAGTTGCG-1 CAAGGGACACCGGAAA-1 CAAGGGACACGTGAGA-1 ## 0 7 0 5 ## CAAGGGAGTAAGAACT-1 CAAGGGATCTTCGTAT-1 CAATACGTCACACCCT-1 CAATCGACAATTGAAG-1 ## 1 4 1 3 ## CAATGACAGGTTGCCC-1 CAATGACCAAATAAGC-1 CAATGACCATATTCGG-1 CACAACACATGACTCA-1 ## 8 3 4 1 ## CACACAACACCCTATC-1 CACAGATTCCTAACAG-1 CACAGGCTCTGTCCGT-1 CACATGAAGAGGATGA-1 ## 2 4 2 4 ## CACATGAGTGCGTTTA-1 CACCAAAGTAATCAGA-1 CACCGTTCATTAGGCT-1 CACCGTTGTCCAGGTC-1 ## 1 6 3 1 ## CACCGTTTCACTCACC-1 CACCGTTTCTAGACCA-1 CACTGGGAGCTACTGT-1 CACTGGGGTAATGTGA-1 ## 3 7 3 4 ## CACTGTCCATTAGGAA-1 CACTTCGGTAAGTTGA-1 CACTTCGTCAGCATTG-1 CAGATACAGGAACGAA-1 ## 8 2 3 2 ## CAGATACGTAGGAGTC-1 CAGATACGTGCACATT-1 CAGATCAAGTTCACTG-1 CAGATCACAAGGTACG-1 ## 2 0 1 6 ## CAGATTGAGCGCCATC-1 CAGCAATGTGAGCAGT-1 CAGCCAGAGCTGGAGT-1 CAGCCAGCACATCCCT-1 ## 0 9 2 4 ## CAGCCAGGTTCAAGGG-1 CAGCCAGTCAGTGGGA-1 CAGCGTGAGCTTGTGT-1 CAGCGTGCACCCATAA-1 ## 5 7 10 4 ## CAGCGTGGTTGCATGT-1 CAGCGTGGTTGCTCGG-1 CAGGCCACACGAAGAC-1 CAGGCCACAGGTCAAG-1 ## 0 11 1 2 ## CAGGGCTAGAGAGCAA-1 CAGGGCTTCATTCACT-1 CAGGTATCATGGCCCA-1 CAGTGCGCACATACGT-1 ## 4 3 4 0 ## CAGTGCGGTCATTCCC-1 CAGTTAGGTAATCAGA-1 CAGTTCCCACCTGAAT-1 CAGTTCCGTGCATGTT-1 ## 6 2 0 3 ## CAGTTCCGTGTTTACG-1 CATAAGCGTATGGAAT-1 CATACAGGTTGGAGAC-1 CATACTTCAAGAGATT-1 ## 0 9 4 0 ## CATACTTGTCACCGCA-1 CATACTTGTCCCTGTT-1 CATAGACAGGGTGGGA-1 CATAGACGTAGATCGG-1 ## 10 3 1 6 ## CATAGACGTCAATGGG-1 CATCAAGCATAGATGA-1 CATCAAGTCGCTATTT-1 CATCCACAGAAGCGCT-1 ## 0 8 0 0 ## CATCCACAGCGTGCTC-1 CATCCGTGTAGGAGGG-1 CATCGCTCAGGGATAC-1 CATCGGGAGAGCAAGA-1 ## 1 5 10 9 ## CATCGGGTCTCGCGTT-1 CATCGTCAGAAGCGCT-1 CATCGTCTCCGGCAGT-1 CATGAGTAGTTCGGTT-1 ## 0 3 1 7 ## CATGAGTTCTTCCGTG-1 CATGCAATCTCGCCTA-1 CATGCCTCACTTGAGT-1 CATGCCTCAGTGGTGA-1 ## 4 4 1 3 ## CATGCCTTCATCACTT-1 CATGCGGTCTAGCAAC-1 CATGCTCGTTGTACGT-1 CATGCTCTCTGAGCAT-1 ## 5 4 7 0 ## CATGGATAGAGTCTGG-1 CATGGATAGGTAGACC-1 CATGGATCATCTTCGC-1 CATGGATGTAGATTGA-1 ## 0 0 0 1 ## CATGGATTCCAGTTCC-1 CATGGTACAAATCAAG-1 CATTCTAGTAGAATAC-1 CATTGAGTCCCGAGTG-1 ## 2 7 1 6 ## CATTGCCCAGATGCGA-1 CATTGTTAGCTTTGTG-1 CATTGTTGTTGCTAGT-1 CATTTCAGTAATGATG-1 ## 0 2 5 4 ## CATTTCAGTCGAAGCA-1 CCAAGCGGTAGCCCTG-1 CCAATGAAGATGCGAC-1 CCAATGAAGGAACTAT-1 ## 4 3 5 10 ## CCAATGATCTGCTGAA-1 CCAATTTGTGTCGCTG-1 CCACAAACATGCGGTC-1 CCACACTGTATCGAGG-1 ## 1 7 7 1 ## CCACCATAGAATTTGG-1 CCACCATAGTATAACG-1 CCACGAGTCGCCTATC-1 CCACGAGTCTTAGCCC-1 ## 6 0 2 5 ## CCACGTTTCCATCAGA-1 CCACGTTTCTATCCAT-1 CCACTTGAGGGTACAC-1 CCACTTGTCACTAGCA-1 ## 0 0 1 1 ## CCACTTGTCTGAGATC-1 CCATCACCAACCGATT-1 CCCAACTTCCCGAAAT-1 CCCGAAGAGACAACAT-1 ## 1 1 0 2 ## CCCGAAGAGTGAGTGC-1 CCCGAAGTCCGATGCG-1 CCCGGAACAACTAGAA-1 CCCGGAACATGTGGTT-1 ## 4 10 3 1 ## CCCTAACAGGATCACG-1 CCCTAACCACAGTCAT-1 CCCTAACCATTAAAGG-1 CCCTAACGTGTTCAGT-1 ## 2 1 9 7 ## CCCTAACTCATCTACT-1 CCCTAACTCTGCGGAC-1 CCCTCAAGTGACACGA-1 CCCTCAATCCCATAAG-1 ## 7 6 8 2 ## CCCTCAATCCCTTCCC-1 CCCTCTCGTGAGATAT-1 CCCTTAGGTCTCTCTG-1 CCCTTAGTCATGGCCG-1 ## 5 10 3 0 ## CCCTTAGTCGTTCATT-1 CCGAACGGTAAGCTCT-1 CCGATCTCACGCGTCA-1 CCGATCTTCAGAGTTC-1 ## 7 0 3 4 ## CCGATGGGTTGGGCCT-1 CCGATGGTCCGGGACT-1 CCGCAAGCAGACCTGC-1 CCGCAAGGTGTTGATC-1 ## 5 4 3 3 ## CCGGACAAGGTTTGAA-1 CCGGGTAAGTGTAGAT-1 CCGGTGAAGGCCACCT-1 CCGGTGACACAACCGC-1 ## 9 9 3 7 ## CCGGTGAGTCAAAGAT-1 CCGTGAGAGATTACCC-1 CCGTGAGCAAGCCTGC-1 CCGTGAGCATCCGCGA-1 ## 2 7 1 2 ## CCGTTCATCAGATTGC-1 CCTAACCTCACTACGA-1 CCTAAGAAGAGGTCGT-1 CCTAAGACAGTAGGAC-1 ## 2 2 8 1 ## CCTAAGACAGTGTGGA-1 CCTAAGACATGGCCCA-1 CCTAAGAGTATTTCCT-1 CCTACGTAGCGAGGAG-1 ## 2 2 9 0 ## CCTACGTAGTGGCCTC-1 CCTACGTCACGCGTCA-1 CCTACGTTCAAACTGC-1 CCTATCGAGTATTGCC-1 ## 2 2 0 1 ## CCTATCGCAGTTGAAA-1 CCTCAACAGATTGGGC-1 CCTCAACTCGCACGAC-1 CCTCAACTCTCTGCTG-1 ## 7 0 5 0 ## CCTCAGTCATCCTATT-1 CCTCAGTCATCGGATT-1 CCTCAGTCATGGGAAC-1 CCTCATGCACTTCATT-1 ## 2 5 4 1 ## CCTCATGTCGTTCCCA-1 CCTCCAACAAGCTCTA-1 CCTCCAACACTGCACG-1 CCTCCAAGTGCCTGCA-1 ## 5 11 0 0 ## CCTCCTCTCACTTGTT-1 CCTCCTCTCATCCTAT-1 CCTCTAGAGAGCCATG-1 CCTCTAGCATGCAGGA-1 ## 0 10 0 8 ## CCTCTCCGTGACCGTC-1 CCTCTCCTCGTGGGAA-1 CCTGCATAGCTAAACA-1 CCTGCATCAGTCGTTA-1 ## 1 5 1 1 ## CCTGCATGTCAGACGA-1 CCTGCATTCGATTCCC-1 CCTGCATTCTCCAATT-1 CCTTCAGCAGCTACTA-1 ## 10 3 2 6 ## CCTTGTGAGCGTGCTC-1 CCTTGTGCAAGCAGGT-1 CCTTGTGCAGATCATC-1 CCTTGTGGTCGCATGC-1 ## 9 5 0 1 ## CCTTGTGTCACCTGGG-1 CCTTGTGTCCACGTCT-1 CCTTGTGTCGTTCCTG-1 CCTTTGGAGAGCAGCT-1 ## 3 6 0 0 ## CCTTTGGAGGGCTTCC-1 CCTTTGGGTCACAGTT-1 CCTTTGGTCAGCTTCC-1 CGAAGGAAGGATATAC-1 ## 7 2 0 1 ## CGAAGGATCGAATGCT-1 CGAATTGAGTGCTCAT-1 CGAATTGCATTCTCTA-1 CGAGAAGGTGACAGCA-1 ## 6 7 6 0 ## CGAGGAAAGGCAGGTT-1 CGAGGAAGTAGTGCGA-1 CGAGGCTGTCCGTACG-1 CGAGTGCAGCGCTTCG-1 ## 0 0 2 3 ## CGAGTGCCAACTAGAA-1 CGAGTGCGTTCGGTTA-1 CGAGTTAAGCGACCCT-1 CGAGTTAAGCGCACAA-1 ## 2 2 9 2 ## CGAGTTAGTGTGTCCG-1 CGAGTTATCATGCCCT-1 CGAGTTATCTGTCGTC-1 CGATCGGAGATGCGAC-1 ## 1 1 6 8 ## CGATCGGTCCCTCAAC-1 CGATCGGTCGGACTTA-1 CGATGCGTCTTAGCAG-1 CGATGGCAGGTCTACT-1 ## 3 0 2 3 ## CGATGGCCACTGGCGT-1 CGCAGGTCACTGGCGT-1 CGCATAAGTTCTCCCA-1 CGCATAATCTCCCATG-1 ## 3 1 6 3 ## CGCATGGAGATGTTAG-1 CGCATGGAGCCTAGGA-1 CGCCAGATCGGTGAAG-1 CGCCATTGTTGGATCT-1 ## 5 8 0 4 ## CGGAACCCAGGCTTGC-1 CGGAACCGTGAGTAAT-1 CGGAACCTCTTAATCC-1 CGGAATTTCTCCTACG-1 ## 5 6 2 3 ## CGGAGAAAGCGCCTTG-1 CGGAGAACAGGACATG-1 CGGGACTTCAAATAGG-1 CGGGCATCACCACATA-1 ## 2 0 0 0 ## CGGGTCATCGTTCTCG-1 CGGGTGTAGTTGAATG-1 CGGTCAGAGTCGGCAA-1 CGGTCAGCAGAGCGTA-1 ## 4 0 4 10 ## CGGTCAGTCATGGCCG-1 CGTAAGTTCAATCTTC-1 CGTAATGGTTAGGCCC-1 CGTAATGTCGGCATCG-1 ## 10 6 4 3 ## CGTAGTACAGTTAGGG-1 CGTCAAACAGTCAACT-1 CGTCAAAGTCATAGTC-1 CGTCCATAGAACCGCA-1 ## 0 3 1 10 ## CGTCCATAGGCATGCA-1 CGTCCATAGTCAGCCC-1 CGTCCATTCACTACGA-1 CGTGAATGTCCATACA-1 ## 3 2 1 5 ## CGTGCTTCACTGGAAG-1 CGTGCTTGTGTCCACG-1 CGTGCTTGTTGGCTAT-1 CGTGTCTCAAAGCTAA-1 ## 0 1 8 0 ## CGTGTCTCAGGTGGAT-1 CGTGTCTCATCTCAAG-1 CGTGTCTGTCACAATC-1 CGTGTCTTCGGCTTGG-1 ## 0 6 0 7 ## CGTTAGATCGAGTCTA-1 CGTTCTGCAGAGTTGG-1 CGTTCTGGTCAGTCCG-1 CGTTCTGGTGGCTCTG-1 ## 5 8 5 1 ## CGTTGGGAGCCGATCC-1 CGTTGGGGTGACCTGC-1 CGTTGGGTCAGATTGC-1 CTAACCCAGTCATTGC-1 ## 2 9 3 6 ## CTAACCCCACGTTGGC-1 CTAACTTAGGGAACAA-1 CTAACTTAGGTATAGT-1 CTAAGTGAGGTAGCAC-1 ## 1 3 4 1 ## CTAAGTGCAGCGCTTG-1 CTAAGTGGTCATGCAT-1 CTAAGTGGTCCTGTTC-1 CTAAGTGGTGGCAACA-1 ## 5 9 3 0 ## CTAAGTGTCGCCAACG-1 CTAAGTGTCGTTGTTT-1 CTACAGAAGTAAGACT-1 CTACAGAGTCTACACA-1 ## 8 0 7 3 ## CTACAGATCAGCACCG-1 CTACATTTCTCTAAGG-1 CTACCTGAGTTGGGAC-1 CTACGGGCAACCGTGC-1 ## 0 2 1 3 ## CTACGGGCATGGACAG-1 CTAGACACATCCTTCG-1 CTAGGTAAGTGAGTTA-1 CTAGGTAGTGATTAGA-1 ## 1 2 2 1 ## CTATAGGCAGCACAGA-1 CTATAGGGTCAGCGTC-1 CTATAGGGTCCCACGA-1 CTATAGGGTTTCGACA-1 ## 0 1 0 3 ## CTATCCGTCGTCTAAG-1 CTCAAGATCAAGCTGT-1 CTCAATTAGATAGCAT-1 CTCAATTGTAGACTGG-1 ## 4 0 5 0 ## CTCAATTTCGTTCTGC-1 CTCACTGCAGTCTTCC-1 CTCACTGTCATCGCTC-1 CTCACTGTCTGACGCG-1 ## 0 0 0 3 ## CTCAGAAGTAATGCTC-1 CTCAGAATCCGTTGGG-1 CTCAGGGAGATAGCAT-1 CTCAGGGGTACCCAGC-1 ## 7 3 3 5 ## CTCAGTCCAACCGCTG-1 CTCATCGCAACATACC-1 CTCATCGCAGCAGTGA-1 CTCATCGGTCGTGCCA-1 ## 1 5 0 0 ## CTCATCGGTTGAATCC-1 CTCATGCGTTAATCGC-1 CTCATTAAGTTTGTCG-1 CTCATTACAACCACGC-1 ## 0 0 0 7 ## CTCATTAGTTCATCTT-1 CTCATTATCAGTAGGG-1 CTCATTATCGGCATTA-1 CTCCAACCACACCTAA-1 ## 1 3 0 1 ## CTCCACATCTCCCATG-1 CTCCATGAGCGTGAAC-1 CTCCCAAGTACTAACC-1 CTCCCAATCTTCGGTC-1 ## 3 3 0 3 ## CTCCCTCAGATCCGAG-1 CTCCCTCCAACTTCTT-1 CTCCCTCCATCTCAAG-1 CTCCGATAGTACCATC-1 ## 0 1 2 7 ## CTCCGATGTAGGGAGG-1 CTCCTCCAGGTTAAAC-1 CTCCTCCCATGACGAG-1 CTCCTTTAGCACCGTC-1 ## 11 0 1 0 ## CTCCTTTCACGGCGTT-1 CTCCTTTGTTTACCTT-1 CTCCTTTTCCTACGGG-1 CTCGAGGCACGTGTGC-1 ## 5 2 7 0 ## CTCGAGGGTCGCAGTC-1 CTCTCAGAGTTTGCTG-1 CTCTCGACAGAACTTC-1 CTCTCGAGTCGTGATT-1 ## 1 0 4 0 ## CTCTCGAGTGACTAAA-1 CTCTGGTAGCCAGAGT-1 CTCTGGTCAGAAACCG-1 CTCTGGTGTGCGTTTA-1 ## 2 1 3 2 ## CTCTGGTTCGATGCAT-1 CTGAATGCACGACCTG-1 CTGAATGCACGCAAAG-1 CTGAGGCGTGTTCGTA-1 ## 0 0 0 0 ## CTGCAGGAGTCTGCAT-1 CTGCAGGGTATGTCAC-1 CTGCATCAGACATAGT-1 CTGCATCTCCCTCTTT-1 ## 3 2 4 3 ## CTGCCATCACGTCATA-1 CTGCCATGTGTCTTAG-1 CTGCCTAAGCTCATAC-1 CTGCCTAAGTTGAAAC-1 ## 3 5 7 2 ## CTGCCTACAGGTCAAG-1 CTGCCTATCTCATAGG-1 CTGCGAGAGGGCCTCT-1 CTGCGAGAGGTCACTT-1 ## 1 4 11 0 ## CTGCGAGAGTGGAAAG-1 CTGGACGAGTCTCGTA-1 CTGGCAGGTAGCACGA-1 CTGGCAGTCTCCCATG-1 ## 1 3 0 3 ## CTGGTCTTCCCAGGCA-1 CTGTACCAGCGCGTTC-1 CTGTACCGTTTCTTAC-1 CTGTAGAAGCACTCCG-1 ## 9 0 0 8 ## CTGTAGAAGGTAGACC-1 CTGTAGAAGGTGCATG-1 CTGTATTCATTCACCC-1 CTGTCGTCAAGAGTAT-1 ## 5 0 1 8 ## CTGTCGTCATATGCGT-1 CTGTCGTGTTGCGGCT-1 CTGTCGTGTTTGACAC-1 CTGTGAAAGGAGTATT-1 ## 0 0 0 11 ## CTGTGAATCTGGCTGG-1 CTGTGGGTCACCGCTT-1 CTTACCGCAAGACCGA-1 CTTACCGCATACATCG-1 ## 11 7 6 4 ## CTTAGGAAGTGGCCTC-1 CTTAGGAGTATACGGG-1 CTTAGGAGTCATCCGG-1 CTTAGGATCCCATAAG-1 ## 10 4 9 5 ## CTTCCGAAGCGGACAT-1 CTTCCTTAGCCAAGGT-1 CTTCCTTTCAGCATTG-1 CTTCGGTAGACCCGCT-1 ## 5 3 3 1 ## CTTCGGTCAGCTGTCG-1 CTTCGGTTCCCTTGTG-1 CTTCTAAAGGCTGTAG-1 CTTCTAATCACCCTGT-1 ## 0 1 1 1 ## CTTCTAATCGACTCCT-1 CTTCTCTAGTAGCATA-1 CTTCTCTGTAAGTAGT-1 CTTCTCTTCATGGCCG-1 ## 5 7 3 4 ## CTTGATTAGAGGCGTT-1 CTTGATTGTGCGCTCA-1 CTTTCAAGTCTGTCAA-1 GAAACCTAGAGAGAAC-1 ## 0 0 2 10 ## GAAACCTGTGTTGACT-1 GAAACCTTCAGGCGAA-1 GAAACCTTCGCAATTG-1 GAAATGAGTGCCTGAC-1 ## 6 2 0 4 ## GAAATGAGTTAGGCCC-1 GAACACTAGTGCCTCG-1 GAACACTCAGCTGTTA-1 GAACGTTAGGGATCGT-1 ## 6 0 2 4 ## GAACGTTGTCGCATTA-1 GAACTGTAGACTCCGC-1 GAACTGTCAATAGTGA-1 GAACTGTGTGTGAATA-1 ## 11 2 2 0 ## GAAGAATTCGGATAAA-1 GAAGCCCGTGACTAAA-1 GAAGCCCTCCGAACGC-1 GAAGCGAAGGTTCTTG-1 ## 4 1 4 1 ## GAAGCGAGTACTGCGC-1 GAAGGACCATGTGGTT-1 GAAGTAACATTACGGT-1 GAAGTAAGTCTGTAGT-1 ## 3 0 1 3 ## GAATAGAGTGGAATGC-1 GAATCACCAACAGCTT-1 GAATCACCATGACTCA-1 GAATCGTAGAACTGAT-1 ## 0 7 0 4 ## GAATCGTTCCGTCACT-1 GACACGCCAGCATGCC-1 GACACGCCATTGACAC-1 GACAGCCGTGTTCATG-1 ## 0 3 0 1 ## GACATCATCCAACCGG-1 GACCAATAGTAAACAC-1 GACCAATCACCCTAAA-1 GACCAATTCGCACTCT-1 ## 3 0 3 0 ## GACCCAGAGTTCCGTA-1 GACCCAGTCGTACACA-1 GACCGTGGTAGATTGA-1 GACCGTGGTAGCTGTT-1 ## 0 5 2 2 ## GACCGTGTCAGTCATG-1 GACCGTGTCTACCTTA-1 GACGCTGGTGGTTCTA-1 GACGCTGGTTCGTGCG-1 ## 4 3 0 4 ## GACGTTAAGCTGAGCA-1 GACTATGCAACAGCTT-1 GACTATGCACAACGAG-1 GACTATGCACAGCCTG-1 ## 8 3 1 0 ## GACTCTCGTAGAGTTA-1 GAGAAATCAGCGATTT-1 GAGAAATTCCAAACCA-1 GAGACCCGTACAGTTC-1 ## 0 10 0 1 ## GAGAGGTCAGGCGTTC-1 GAGATGGTCGCCGATG-1 GAGCCTGTCGCCTATC-1 GAGCTGCCACTAGGCC-1 ## 1 2 1 8 ## GAGCTGCTCCGTGCGA-1 GAGGCAACAAGTGGCA-1 GAGGCAACATCCTATT-1 GAGGCCTTCCGCAACG-1 ## 3 0 1 3 ## GAGGGATAGGTGATAT-1 GAGGGATCACTGGATT-1 GAGGGATGTCTAACTG-1 GAGGGTACATGGATCT-1 ## 1 9 2 6 ## GAGGGTAGTGTTGACT-1 GAGGGTAGTTAGGGAC-1 GAGTCTACAGCGCTTG-1 GAGTGAGGTGGCACTC-1 ## 3 5 2 5 ## GAGTGAGTCTTACGGA-1 GAGTGTTTCATTGCCC-1 GAGTGTTTCATTGCGA-1 GAGTTACTCATACGAC-1 ## 1 0 9 9 ## GAGTTACTCGTAGGAG-1 GAGTTGTAGGCTCACC-1 GAGTTGTCAAGTATAG-1 GAGTTGTCACCCAACG-1 ## 8 2 3 5 ## GAGTTGTGTCTGCATA-1 GAGTTGTTCGCCGAAC-1 GAGTTTGCACAATGAA-1 GAGTTTGGTAGTCGGA-1 ## 6 2 0 0 ## GAGTTTGTCGACATCA-1 GATAGAAGTAGAGGAA-1 GATCACATCCTCAGGG-1 GATCACATCTGAACGT-1 ## 8 9 9 10 ## GATCAGTCATCACGGC-1 GATCCCTAGTCACTCA-1 GATCCCTCAATCAAGA-1 GATCGTAAGTAGCCAG-1 ## 4 3 2 3 ## GATCGTATCTAATTCC-1 GATGAGGCACAAGGTG-1 GATGAGGGTAATCAGA-1 GATGAGGTCATGTCTT-1 ## 4 1 7 4 ## GATGAGGTCTGAGTCA-1 GATGATCCATATCTCT-1 GATGATCGTTAGGAGC-1 GATGATCTCTCCCATG-1 ## 2 2 2 3 ## GATGCTAAGAAGCGAA-1 GATGCTAGTCTCGGGT-1 GATGCTATCGCTGTCT-1 GATGGAGCACAACGCC-1 ## 8 5 4 9 ## GATGGAGTCGTTAGAC-1 GATGTTGCACGATTCA-1 GATTCGACAGGCGAAT-1 GATTCGATCCCTGGTT-1 ## 7 0 0 4 ## GATTCGATCTTACCAT-1 GATTCTTGTTAGCTAC-1 GATTCTTTCCGAGTGC-1 GATTCTTTCCTTCACG-1 ## 3 7 5 1 ## GATTGGTGTAGTTCCA-1 GATTGGTTCTCCCTAG-1 GATTTCTAGTCGAAAT-1 GATTTCTCAATAGAGT-1 ## 1 11 5 7 ## GATTTCTTCCCATGGG-1 GCAACATAGGTAAAGG-1 GCAACATCAAATAGCA-1 GCACATATCCAGTTCC-1 ## 3 0 2 0 ## GCACGGTCACGAAAGC-1 GCACGGTCAGGAGACT-1 GCACGTGCAAGTGCTT-1 GCACGTGTCTCTGACC-1 ## 1 4 0 8 ## GCACGTGTCTGTCTCG-1 GCACGTGTCTTGGATG-1 GCACTAAGTCGTCTCT-1 GCACTAAGTGTGCTTA-1 ## 2 3 5 5 ## GCACTAAGTTCGGTAT-1 GCAGCCAAGTCGAAAT-1 GCAGCCACAGCCCAGT-1 GCAGCTGCAGCGAACA-1 ## 0 8 0 1 ## GCAGCTGGTGCCCAGT-1 GCAGGCTGTCGGCCTA-1 GCAGGCTGTTAGAAAC-1 GCAGTTATCTGCACCT-1 ## 1 3 4 5 ## GCATCGGAGCTAGAAT-1 GCATCGGCAGTAGGAC-1 GCATCGGGTAACATGA-1 GCATCGGTCAGGAAAT-1 ## 10 5 3 8 ## GCATCGGTCTGCGAGC-1 GCATCTCCATCTATCT-1 GCATCTCGTCTACAAC-1 GCATGATCATTACTCT-1 ## 0 2 1 1 ## GCATGATGTACATACC-1 GCATTAGCAAGACGAC-1 GCATTAGGTCTGGTTA-1 GCATTAGTCGGCATAT-1 ## 0 6 5 0 ## GCCAACGTCTCTCGCA-1 GCCAGGTGTTTCCAAG-1 GCCAGTGAGGCACTAG-1 GCCAGTGAGTGCCTCG-1 ## 10 4 11 3 ## GCCATTCAGATGACCG-1 GCCATTCCAACAGAGC-1 GCCATTCCATTCAGGT-1 GCCATTCTCAGGCGAA-1 ## 4 8 1 0 ## GCCCAGAAGCGGGTAT-1 GCCCAGAGTGCTTATG-1 GCCCAGATCAGGGTAG-1 GCCCAGATCCTGTAGA-1 ## 0 2 1 1 ## GCCCGAACATACCACA-1 GCCCGAAGTTGTGCCG-1 GCCCGAATCTATTTCG-1 GCCGTGATCCACGTCT-1 ## 9 1 2 6 ## GCCTGTTCACGGAAGT-1 GCGAGAATCCCTTGTG-1 GCGGAAACATGGGTCC-1 GCGGAAATCAGACCTA-1 ## 4 1 7 2 ## GCGGATCTCTCCCATG-1 GCGGATCTCTGCACCT-1 GCGTGCAAGCCAAGGT-1 GCGTGCATCAACGCTA-1 ## 3 6 5 0 ## GCGTTTCGTGGTAATA-1 GCTACAAAGAACTGAT-1 GCTACCTCAAGTATAG-1 GCTACCTCATCGATCA-1 ## 8 6 0 2 ## GCTACCTTCCCTCATG-1 GCTCAAATCACACCCT-1 GCTCAAATCAGTGTTG-1 GCTCAAATCCGTGGTG-1 ## 6 3 3 1 ## GCTGAATAGATTAGTG-1 GCTGAATGTAGTTACC-1 GCTGCAGCATCATCCC-1 GCTGCAGGTCTTGTCC-1 ## 3 10 6 5 ## GCTGCAGTCCCAGGAC-1 GCTGGGTGTCAACGCC-1 GCTGGGTTCAGTGTGT-1 GCTTCACAGCTTAAGA-1 ## 3 0 4 5 ## GCTTCACCATTGTGCA-1 GCTTGGGGTATAGGGC-1 GCTTGGGGTCTAACGT-1 GCTTGGGGTGATGAAT-1 ## 8 3 0 2 ## GCTTTCGGTCATCCGG-1 GGAAGTGAGCTGAGCA-1 GGAAGTGAGGGACTGT-1 GGAAGTGCATGGGAAC-1 ## 3 7 4 3 ## GGAAGTGGTAGAGCTG-1 GGAATCTGTCATCGGC-1 GGAATGGAGATCACCT-1 GGAATGGAGGATACCG-1 ## 2 9 8 6 ## GGAATGGAGTCGCCCA-1 GGACGTCCACAACGAG-1 GGACGTCCACTTGGCG-1 GGACGTCTCTTGGTGA-1 ## 4 4 0 1 ## GGAGAACCAAGAGTGC-1 GGAGATGTCCTCTAGC-1 GGAGCAAAGCCTGTCG-1 GGAGGATAGGGCAACT-1 ## 8 0 7 1 ## GGAGGATCAGGTCTCG-1 GGAGGTAGTTGCAAGG-1 GGAGGTATCAAGAGGC-1 GGATCTATCGGTTGTA-1 ## 3 1 0 3 ## GGATGTTAGCGGACAT-1 GGATGTTGTGCCGTTG-1 GGATGTTTCAGGTGTT-1 GGCACGTAGCACAAAT-1 ## 4 4 0 1 ## GGCACGTAGGATCACG-1 GGCACGTCAGTCTGGC-1 GGCACGTTCGCAGTGC-1 GGCTGTGTCTTAAGGC-1 ## 2 1 4 9 ## GGCTTGGAGATAGTCA-1 GGCTTGGAGGCATCAG-1 GGCTTGGAGTAACCTC-1 GGCTTGGCATCGCTCT-1 ## 4 6 0 6 ## GGCTTGGGTGTCATGT-1 GGCTTTCAGAAGGCTC-1 GGGAAGTGTTGCACGC-1 GGGACCTAGTTTGCTG-1 ## 1 10 1 0 ## GGGACCTGTGAGATCG-1 GGGACCTTCCTGGGAC-1 GGGACTCGTTAAGGGC-1 GGGAGATCACAATGTC-1 ## 0 11 5 6 ## GGGAGATGTGGCTCTG-1 GGGAGATTCAGGGTAG-1 GGGAGATTCAGTGTTG-1 GGGATCCAGGTTAAAC-1 ## 0 6 4 2 ## GGGATGAGTGGATCGA-1 GGGCCATGTCTTGCGG-1 GGGCGTTAGATCCGAG-1 GGGCGTTGTACAGTCT-1 ## 6 0 4 1 ## GGGCGTTGTTTACCAG-1 GGGCTACTCGTGCAGC-1 GGGCTCACACTGTGTA-1 GGGCTCACAGAGTGAC-1 ## 6 0 3 9 ## GGGTAGACATTGCCTC-1 GGGTATTCACTCCTGT-1 GGGTATTGTATCACCA-1 GGGTATTGTGTATCCA-1 ## 1 5 0 6 ## GGGTCACGTAACGATA-1 GGGTCACGTGCGTCGT-1 GGGTCTGAGCTAGAAT-1 GGGTGTCAGTAGACCG-1 ## 4 5 5 4 ## GGGTGTCGTGCAGGAT-1 GGGTTATCAGACCTAT-1 GGGTTATTCCTGTAGA-1 GGGTTTATCATGGTAC-1 ## 1 5 4 7 ## GGTAACTAGAGCCGTA-1 GGTAACTCAACGCATT-1 GGTAACTCAATCAAGA-1 GGTAATCCAGAACCGA-1 ## 1 0 5 3 ## GGTAATCCAGGCTATT-1 GGTAATCGTTGCATTG-1 GGTAATCGTTTACACG-1 GGTAGAGCAACGGTAG-1 ## 5 1 0 6 ## GGTCACGAGCGATGGT-1 GGTCACGGTGGACAGT-1 GGTCACGTCATCCCGT-1 GGTCACGTCGGACTGC-1 ## 7 0 5 3 ## GGTGATTAGCCATGCC-1 GGTGTTATCCATGATG-1 GGTTAACCACGCACCA-1 GGTTAACTCGTGTTCC-1 ## 2 0 1 0 ## GGTTCTCCAAGGCGTA-1 GTAACACAGACGAGCT-1 GTAACACAGCATTTGC-1 GTAACACAGTTTGAGA-1 ## 4 10 1 8 ## GTAACCATCTTCCTAA-1 GTAAGTCCACCTGCAG-1 GTAAGTCCAGCTGCCA-1 GTAAGTCTCAGCATTG-1 ## 0 1 4 4 ## GTAATGCAGTCTCGTA-1 GTAATGCCACAGCTTA-1 GTAATGCTCAAAGGTA-1 GTAATGCTCTCAATCT-1 ## 10 0 0 0 ## GTACAACCAGGCACAA-1 GTACAGTCAGCAGACA-1 GTACAGTTCAATCCGA-1 GTACAGTTCGAGCTGC-1 ## 0 0 1 1 ## GTAGAGGAGTCATAGA-1 GTAGGAGAGTGCTCAT-1 GTAGGAGGTACGAGTG-1 GTAGGAGGTGAACGGT-1 ## 2 5 4 2 ## GTAGGAGTCGTAGGAG-1 GTAGTACAGCGTTCAT-1 GTAGTACTCTGGTTGA-1 GTATTTCGTAGTCCTA-1 ## 6 3 2 2 ## GTCAAACAGCATCAGG-1 GTCAAGTCACTTGTCC-1 GTCAAGTCATGTGCCG-1 GTCAAGTTCAAAGACA-1 ## 0 1 0 0 ## GTCACGGAGATCCCAT-1 GTCACTCAGAGCAAGA-1 GTCACTCAGGGTTAGC-1 GTCACTCCAGAGTGTG-1 ## 1 4 0 2 ## GTCAGCGTCAGCTGAT-1 GTCATCCTCGTGGTAT-1 GTCATGAAGAGCAAGA-1 GTCATGAAGTCGGCAA-1 ## 4 0 5 1 ## GTCATGACACTCTGCT-1 GTCATGAGTTAGGCCC-1 GTCCACTCACTGGAAG-1 GTCCTCACACAACCGC-1 ## 4 0 2 0 ## GTCCTCATCTACACAG-1 GTCGAATAGGGTTAGC-1 GTCGAATCACTTGGGC-1 GTCGAATGTGGAAATT-1 ## 11 7 2 9 ## GTCGAATTCATTACGG-1 GTCGCGATCCGACATA-1 GTCGTAAAGGTCGCCT-1 GTCGTAAGTGCAGGAT-1 ## 2 9 0 2 ## GTCTACCAGTTGCGCC-1 GTCTACCGTTTACACG-1 GTCTACCTCAGTCATG-1 GTCTACCTCTACCCAC-1 ## 2 5 4 2 ## GTCTAGATCATCACTT-1 GTCTCACCATGCCATA-1 GTCTCACGTCACAGTT-1 GTCTTTAAGGCTAGCA-1 ## 1 11 1 6 ## GTCTTTACACAGAAGC-1 GTGAGCCAGTGGGAAA-1 GTGAGCCGTCGCATTA-1 GTGAGGAGTTTGTTGG-1 ## 0 7 3 1 ## GTGAGGATCAACCTCC-1 GTGAGGATCGCTTGCT-1 GTGAGTTGTCATCGGC-1 GTGATGTAGACCAAGC-1 ## 0 3 0 8 ## GTGATGTTCGACATTG-1 GTGCACGCACCGTGAC-1 GTGCGTGAGCTCCCTT-1 GTGCGTGTCCTAACAG-1 ## 1 8 0 0 ## GTGCTGGTCTCAAAGC-1 GTGCTTCCACGGGTAA-1 GTGCTTCGTGCATACT-1 GTGCTTCTCCGATAAC-1 ## 0 4 1 0 ## GTGGCGTGTCATACCA-1 GTGGGAAGTCTTGCGG-1 GTGGTTATCGTGTGGC-1 GTGTAACGTGCCAAGA-1 ## 4 0 3 3 ## GTGTAACGTTATGTCG-1 GTGTAACTCTGGTGGC-1 GTGTCCTAGAGATGCC-1 GTGTGATAGACATCCT-1 ## 0 0 4 6 ## GTGTGATCACGGATCC-1 GTGTGGCCATACAGAA-1 GTGTGGCCATATCTCT-1 GTGTGGCGTCCTCCTA-1 ## 8 7 3 0 ## GTGTTAGCACAATTCG-1 GTGTTAGCACGTAGTT-1 GTGTTAGCAGAAATCA-1 GTGTTAGCATGTCAGT-1 ## 4 2 0 8 ## GTGTTAGTCGGACTTA-1 GTGTTCCAGTAGCAAT-1 GTGTTCCGTCCGGTCA-1 GTGTTCCTCCGCCTAT-1 ## 0 3 3 1 ## GTGTTCCTCCTCTAAT-1 GTTACAGCAGTCGGAA-1 GTTACAGTCTGATTCT-1 GTTACCCAGTTCGCAT-1 ## 0 8 8 0 ## GTTACCCTCCACAAGT-1 GTTACGAAGCGCACAA-1 GTTACGATCAATCCGA-1 GTTACGATCTACCTTA-1 ## 8 0 0 1 ## GTTAGACGTCACCACG-1 GTTAGACTCATTTGCT-1 GTTAGTGCAAATAAGC-1 GTTAGTGGTTACCCAA-1 ## 5 6 7 7 ## GTTAGTGTCCCGAACG-1 GTTCATTGTAACCCGC-1 GTTCATTTCCCTCGAT-1 GTTCCGTAGCGTCGAA-1 ## 1 1 5 0 ## GTTCGCTCATGACTGT-1 GTTCGCTGTACCAGAG-1 GTTCTATGTAGAATAC-1 GTTGCGGAGGAAAGGT-1 ## 4 1 0 6 ## GTTGCTCAGACATAAC-1 GTTGTAGTCCGCTTAC-1 GTTGTCCCACTGAGGA-1 GTTGTCCGTACTAGCT-1 ## 6 10 11 2 ## GTTGTCCGTCGCACGT-1 GTTGTGAAGCGGTAAC-1 GTTGTGAAGTCAAGCG-1 GTTGTGATCGAGATGG-1 ## 7 4 2 0 ## GTTGTGATCGTCAACA-1 GTTTACTTCAAGTGGG-1 GTTTGGAAGATGATTG-1 GTTTGGACAGCTTCGG-1 ## 0 2 0 3 ## TAACACGAGTACTGGG-1 TAACACGCATTCGGGC-1 TAACACGGTCAAAGTA-1 TAACACGGTCGTCTCT-1 ## 2 1 1 8 ## TAACTTCTCCGTGTAA-1 TAAGCACAGCATTGAA-1 TAAGCCACAACCAATC-1 TAAGTCGAGTCAGCGA-1 ## 5 1 9 1 ## TAATTCCCAGGGTTGA-1 TAATTCCGTTCTCCCA-1 TAATTCCTCCAAGCAT-1 TACAACGTCTCATTAC-1 ## 7 6 5 0 ## TACACCCGTTCTCCCA-1 TACATTCAGCTGAGCA-1 TACATTCGTTGTCCCT-1 TACCCACAGAAACTAC-1 ## 4 1 1 2 ## TACCCACGTCAAGCGA-1 TACCCACTCTGCGATA-1 TACCCGTAGCAAGGAA-1 TACCCGTCACTCAGAT-1 ## 2 1 6 10 ## TACCCGTCACTGCGAC-1 TACCCGTGTACAGAAT-1 TACCCGTGTTGCATAC-1 TACCCGTTCTGCGATA-1 ## 4 3 0 0 ## TACCGAAAGGTGCATG-1 TACCGAATCTAAACGC-1 TACCTCGGTGATTAGA-1 TACCTCGTCCCAACTC-1 ## 0 0 7 7 ## TACCTGCCACACTTAG-1 TACCTGCGTAAGACCG-1 TACGCTCGTAGTCGGA-1 TACGCTCGTTACCCAA-1 ## 2 11 3 5 ## TACGGGCCAGGTGTGA-1 TACGGGCTCAACCTCC-1 TACGGTATCATACGGT-1 TACGGTATCGCTTGCT-1 ## 1 0 10 8 ## TACGTCCAGAAACCAT-1 TACGTCCAGCTCCATA-1 TACGTCCGTCAGGTGA-1 TACTGCCGTTTGGGAG-1 ## 0 1 8 2 ## TACTTACAGGCCCAAA-1 TACTTACAGGGTAGCT-1 TACTTACTCTTCTGGC-1 TACTTCATCGGACTGC-1 ## 1 8 0 3 ## TACTTGTAGGGCAGTT-1 TAGACCATCCCGTTGT-1 TAGACTGCATAAGCGG-1 TAGATCGGTACATACC-1 ## 1 2 6 4 ## TAGATCGGTCATAAAG-1 TAGCACAAGACCAACG-1 TAGGAGGAGATTTGCC-1 TAGGGTTAGAACGCGT-1 ## 7 5 4 8 ## TAGGGTTAGACTTAAG-1 TAGGGTTAGAGAGAAC-1 TAGGGTTAGTATAGAC-1 TAGGGTTTCGGCCCAA-1 ## 6 4 9 3 ## TAGGTACCATAATCGC-1 TAGGTTGAGGACCCAA-1 TAGGTTGCAAATCGGG-1 TAGGTTGTCAGTGTTG-1 ## 1 2 0 2 ## TATACCTCACATGTTG-1 TATACCTCAGCTCATA-1 TATACCTTCCATCCGT-1 TATATCCAGCCTCAAT-1 ## 0 4 5 10 ## TATATCCCATCATGAC-1 TATCAGGCAGATAAAC-1 TATCCTAAGCCTCAGC-1 TATCCTACACGACAGA-1 ## 2 3 0 6 ## TATCCTACAGACAAGC-1 TATCGCCCAAATGATG-1 TATCGCCTCCACCTCA-1 TATCTGTAGGGACACT-1 ## 7 2 6 0 ## TATCTGTAGTACAGCG-1 TATCTTGAGCCTCCAG-1 TATCTTGCAGCCTATA-1 TATGTTCTCATTGTGG-1 ## 7 0 1 11 ## TATTCCACATGTGACT-1 TATTGCTGTCACTGAT-1 TATTGCTTCTTCACGC-1 TATTTCGTCGCAGTGC-1 ## 5 0 3 7 ## TATTTCGTCGCTGATA-1 TCAAGACAGACTCCGC-1 TCAAGACAGTGCCGAA-1 TCAAGACAGTTGCATC-1 ## 4 11 8 6 ## TCAAGACGTGGCGTAA-1 TCAAGCAAGGACGCTA-1 TCAATCTAGACCAAAT-1 TCAATCTAGGCAGGTT-1 ## 0 9 5 0 ## TCAATTCAGGTAGTAT-1 TCAATTCCACACAGAG-1 TCACACCAGACTCTAC-1 TCACACCAGCCTCACG-1 ## 1 2 4 4 ## TCACACCCAACATACC-1 TCACACCCAGCTGGTC-1 TCACGCTAGAAACTCA-1 TCACGCTAGTAGTCTC-1 ## 8 6 0 1 ## TCACGCTCAGCTTCGG-1 TCACGGGTCTACGCAA-1 TCACTATCACAACCGC-1 TCACTATCAGCTTCCT-1 ## 1 5 2 2 ## TCACTATGTTCAAAGA-1 TCACTCGCAAGTGGGT-1 TCACTCGGTATCCCTC-1 TCACTCGGTCTCAAGT-1 ## 1 2 3 1 ## TCACTCGTCTGATGGT-1 TCAGCAAAGCGGCTCT-1 TCAGCAAAGGACACTG-1 TCAGCCTGTAGAGCTG-1 ## 4 0 4 7 ## TCAGCCTGTATGGTTC-1 TCAGGGCTCGCAACAT-1 TCAGGTACAAATGGAT-1 TCAGTCCAGAAGCGAA-1 ## 5 4 5 7 ## TCAGTGAAGTACCATC-1 TCAGTGACACAAGCTT-1 TCAGTGACATGACCCG-1 TCAGTTTGTAGACACG-1 ## 2 1 1 5 ## TCATACTTCCTTGAAG-1 TCATCATTCATGAGGG-1 TCATCATTCGCACTCT-1 TCATCCGCACTCTAGA-1 ## 7 6 0 6 ## TCATGCCGTGGCTGCT-1 TCATGTTCAACCTATG-1 TCATTACAGGGAGGAC-1 TCATTACGTTGGGAAC-1 ## 2 9 0 1 ## TCATTGTGTGGCAACA-1 TCCACCAAGGGACCAT-1 TCCACCAGTTGATGTC-1 TCCACCATCTCATTGT-1 ## 2 6 5 1 ## TCCAGAAAGCTCGACC-1 TCCAGAACACAACGAG-1 TCCATCGGTCACCACG-1 TCCATGCGTGGAAGTC-1 ## 5 0 6 6 ## TCCCAGTCATTCTGTT-1 TCCCAGTGTCACGCTG-1 TCCCATGCAACTGATC-1 TCCGAAAGTTGGGACA-1 ## 0 8 1 1 ## TCCGATCCAAGAGGCT-1 TCCGATCGTTACCCAA-1 TCCGATCTCCCTCATG-1 TCCGGGAAGAGAGGGC-1 ## 6 0 2 0 ## TCCGGGATCGCGTGCA-1 TCCGTGTCATGACTGT-1 TCCTCGACATTGAGGG-1 TCCTCTTGTTCTTGTT-1 ## 2 5 0 9 ## TCCTGCACATGTGACT-1 TCCTGCAGTAATGTGA-1 TCCTGCATCAATGCAC-1 TCCTGCATCAGTCAGT-1 ## 3 0 4 0 ## TCCTGCATCTCACGAA-1 TCCTTCTGTACGAAAT-1 TCCTTCTGTGTGTCCG-1 TCCTTCTGTTATCTGG-1 ## 5 6 2 0 ## TCCTTCTTCAGCTCTC-1 TCCTTCTTCCTAACAG-1 TCGAACAAGATAGGGA-1 TCGAACACATGTGACT-1 ## 0 5 5 5 ## TCGAACAGTCCGGACT-1 TCGAAGTAGCGACTAG-1 TCGACCTTCGGCCCAA-1 TCGATTTCAGTGGTGA-1 ## 2 6 3 2 ## TCGATTTTCATCGGGC-1 TCGCACTGTATCTCGA-1 TCGCACTTCACGGAGA-1 TCGCACTTCATTATCC-1 ## 1 2 2 1 ## TCGCAGGTCCAAGGGA-1 TCGCAGGTCGGATAAA-1 TCGCAGGTCTACGGGC-1 TCGCTCAGTCTTTCTA-1 ## 3 1 6 4 ## TCGGATACAATGCAGG-1 TCGGATACATAATCCG-1 TCGGGACTCCTCTTTC-1 TCGGGCACAGCTTCCT-1 ## 2 6 5 0 ## TCGGGCACAGTTGTTG-1 TCGGGCAGTCGACTTA-1 TCGGGCAGTCGATTCA-1 TCGGGTGAGGGCAAGG-1 ## 3 3 6 4 ## TCGGGTGTCATGCCAA-1 TCGGGTGTCCAGTACA-1 TCGGTCTAGAAGGTAG-1 TCGGTCTGTCGCGGTT-1 ## 5 5 2 0 ## TCGGTCTGTTCGTGCG-1 TCGTAGAAGTCAGCCC-1 TCGTAGACAAGACAAT-1 TCGTAGAGTCCTGAAT-1 ## 2 1 6 0 ## TCGTCCAAGCATGGGT-1 TCGTGCTCAATCGAAA-1 TCGTGGGTCTCCACTG-1 TCTAACTAGCACTTTG-1 ## 1 4 3 2 ## TCTAACTCACACAGAG-1 TCTACATGTCAACCTA-1 TCTACATTCCACCCTA-1 TCTACCGAGTGCCTCG-1 ## 5 7 0 1 ## TCTACCGCAAAGCTCT-1 TCTACCGTCAGTGTCA-1 TCTATACAGAGCTGCA-1 TCTATACCAATAACGA-1 ## 3 0 1 0 ## TCTATACCAGCACAAG-1 TCTATCAGTGTCATGT-1 TCTCAGCAGCATACTC-1 TCTCAGCCAGCACACC-1 ## 5 4 6 6 ## TCTCCGACACATTCTT-1 TCTCCGATCCTAGAGT-1 TCTCTGGGTAGGCAGT-1 TCTCTGGTCTACACAG-1 ## 0 1 1 8 ## TCTGCCAAGCACCCAC-1 TCTGCCAAGGAAACGA-1 TCTGCCAAGTCTTCCC-1 TCTGCCATCGTCGATA-1 ## 5 7 10 0 ## TCTGCCATCTTCGTAT-1 TCTGGCTAGCGAGGAG-1 TCTGGCTTCGTGACTA-1 TCTTAGTCAAGCTGTT-1 ## 2 0 4 5 ## TCTTAGTTCCAGCCTT-1 TCTTAGTTCTGCGATA-1 TCTTCCTAGACATACA-1 TCTTCCTAGGTATTGA-1 ## 0 0 5 3 ## TCTTCCTTCGTGGTAT-1 TCTTGCGAGAAGATCT-1 TCTTGCGTCAAAGCCT-1 TCTTGCGTCAAGGCTT-1 ## 7 5 0 5 ## TCTTGCGTCTCCTGAC-1 TCTTTGACACCGGAAA-1 TCTTTGATCCATATGG-1 TGAACGTGTGTGGACA-1 ## 4 3 4 3 ## TGAATCGAGGGCCCTT-1 TGAATCGGTCCATACA-1 TGAATGCCAAGCACCC-1 TGAATGCTCAGCAGAG-1 ## 0 1 4 6 ## TGACAGTAGAGTCGAC-1 TGACAGTTCAACACGT-1 TGACCCTAGGTGAGAA-1 TGACGCGAGTGCCGAA-1 ## 3 3 2 7 ## TGACGCGGTATCGGTT-1 TGACTCCCATCAGCAT-1 TGACTCCTCACGTAGT-1 TGAGACTGTACCCGAC-1 ## 2 2 3 7 ## TGAGACTTCTCTTCAA-1 TGAGCATGTGGGTTGA-1 TGAGCATTCTGCATAG-1 TGAGCGCAGAATCGTA-1 ## 10 2 7 1 ## TGAGCGCAGTCTGCGC-1 TGAGGGAAGATACAGT-1 TGAGGGAAGATAGCTA-1 TGAGGGAAGGTTTGAA-1 ## 7 4 8 1 ## TGAGGGACATCTAACG-1 TGAGGGACATGGCACC-1 TGAGGGATCTGTTGGA-1 TGAGGTTGTCAACGCC-1 ## 4 4 0 0 ## TGAGTCATCTAGAACC-1 TGATCAGAGCGATGGT-1 TGATCAGAGGGAGGGT-1 TGATCAGCAACACACT-1 ## 3 7 3 7 ## TGATCAGGTGGCTCTG-1 TGATCTTAGCTATCTG-1 TGATCTTCAGATTAAG-1 TGATCTTGTAGGAGGG-1 ## 8 11 7 3 ## TGATCTTGTGCCTACG-1 TGATCTTTCTCGCTCA-1 TGATGGTAGAAACCCG-1 TGATGGTAGGGAGGGT-1 ## 3 8 1 2 ## TGATTCTCATGCAGGA-1 TGATTTCAGAAGTGTT-1 TGATTTCGTAGGTAGC-1 TGATTTCTCATGCATG-1 ## 4 1 8 6 ## TGATTTCTCGTAATGC-1 TGCAGGCCAATAGGAT-1 TGCAGGCCATGACTTG-1 TGCATCCAGAAATTCG-1 ## 3 0 1 7 ## TGCATCCAGACTCATC-1 TGCATCCCAATCTCGA-1 TGCATGATCTCTGCTG-1 TGCCGAGCAGTCGAGA-1 ## 0 3 5 3 ## TGCGACGAGCATCCCG-1 TGCGACGCACGGATCC-1 TGCGACGCAGAACCGA-1 TGCGATATCGGCGATC-1 ## 1 1 4 2 ## TGCGGGTGTATGTCTG-1 TGCGGGTGTCAAAGTA-1 TGCTCCAAGTCGAAGC-1 TGCTCCAGTGTCCAAT-1 ## 6 2 6 2 ## TGCTCGTCAACCGTAT-1 TGCTCGTCAATTCTTC-1 TGCTCGTTCAGCTTCC-1 TGCTCGTTCGCTTGAA-1 ## 5 10 6 1 ## TGCTCGTTCGGACCAC-1 TGCTGAATCGGAATGG-1 TGCTTCGAGAACTTCC-1 TGCTTCGGTTCAAGTC-1 ## 2 3 4 4 ## TGCTTGCCAACTTCTT-1 TGGAACTAGGTGAGCT-1 TGGAACTCACGAGAAC-1 TGGAACTTCCCGGTAG-1 ## 3 5 4 6 ## TGGAACTTCTAACACG-1 TGGAGAGCAACGATCT-1 TGGAGGAAGAAATGGG-1 TGGATCAAGCTGTCCG-1 ## 1 0 0 5 ## TGGATCAAGTAGGGTC-1 TGGATCAAGTTGAAGT-1 TGGATCACACTGTGAT-1 TGGATGTCACACCAGC-1 ## 2 0 6 3 ## TGGATGTTCGTGAGAG-1 TGGCGTGTCGCTGTTC-1 TGGGAGACACGCGCTA-1 TGGGAGAGTAGCACAG-1 ## 2 2 4 1 ## TGGGAGAGTATGGTTC-1 TGGGATTAGTGGTCAG-1 TGGGATTTCATGGCCG-1 TGGGCGTCATAATCGC-1 ## 1 2 0 2 ## TGGGCGTGTGCCTATA-1 TGGGTTATCATTCGTT-1 TGGTACAAGATGGGCT-1 TGGTACATCCTTCTAA-1 ## 4 0 0 7 ## TGGTAGTGTGCATCTA-1 TGGTAGTTCTCCGAAA-1 TGGTTAGAGTTGCGCC-1 TGGTTAGGTAACTTCG-1 ## 1 0 6 0 ## TGTAAGCTCACCTCGT-1 TGTACAGAGCATATGA-1 TGTAGACAGAGAAGGT-1 TGTAGACCAATAGTAG-1 ## 7 8 0 1 ## TGTAGACCATGACACT-1 TGTCAGAAGAACAGGA-1 TGTCAGAAGTAGGTTA-1 TGTCAGAGTGAGTTTC-1 ## 8 0 0 0 ## TGTCAGATCCACATAG-1 TGTCCACCAGGCTATT-1 TGTCCACTCAGTCATG-1 TGTCCCACATCACCAA-1 ## 7 7 0 2 ## TGTCCCAGTACGATCT-1 TGTCCTGGTGAAAGTT-1 TGTCCTGTCTGATGGT-1 TGTGAGTAGATGGCGT-1 ## 4 0 0 5 ## TGTGAGTAGCAACTTC-1 TGTGAGTTCTCATGCC-1 TGTGATGCACTGCTTC-1 TGTGATGTCGAGGCAA-1 ## 3 2 6 9 ## TGTGCGGGTATGAAGT-1 TGTGCGGTCACACCCT-1 TGTGCGGTCTGGACTA-1 TGTGCGGTCTTGAACG-1 ## 5 0 3 6 ## TGTGGCGAGCACTAGG-1 TGTGGCGAGCCTAACT-1 TGTGGCGAGGTATCTC-1 TGTGGCGCAATCGCCG-1 ## 1 4 0 0 ## TGTGGCGGTCATCCCT-1 TGTGGCGGTCGTACTA-1 TGTGGCGGTGAGTTTC-1 TGTGGCGGTTAAGGAT-1 ## 3 1 8 5 ## TGTGGCGTCCACCTGT-1 TGTTACTTCCGTCCTA-1 TGTTCATCACAGCTTA-1 TGTTCATGTAGACGTG-1 ## 5 1 4 0 ## TGTTCATGTATCCCAA-1 TGTTCATGTTGTGGAG-1 TGTTCCGAGACCATAA-1 TGTTCTACAACCACAT-1 ## 1 5 9 2 ## TGTTCTACAAGAGCTG-1 TGTTCTATCTTTACAC-1 TGTTGAGAGTGCAGCA-1 TGTTGAGTCTACACTT-1 ## 0 8 0 1 ## TGTTGAGTCTCTCCGA-1 TGTTGGAAGTTAGTAG-1 TGTTGGAGTACGCGTC-1 TGTTGGAGTTATTCCT-1 ## 3 5 6 6 ## TGTTTGTAGGACGCAT-1 TTAATCCAGCGTCGAA-1 TTACAGGCAGCAGTTT-1 TTACCGCTCGGTCATA-1 ## 1 7 11 3 ## TTACGCCAGGGACTGT-1 TTACGCCCACCGGAAA-1 TTACGCCGTCGGTGTC-1 TTACGCCTCTAAACGC-1 ## 6 8 2 3 ## TTACGTTCACGTATAC-1 TTAGGCACATCGTCCT-1 TTAGGCATCCATTGCC-1 TTAGGGTCATCTCAAG-1 ## 4 1 2 0 ## TTAGGGTGTATCGAGG-1 TTAGGGTTCTAGCCAA-1 TTAGTCTAGAACTTCC-1 TTAGTCTGTACACTCA-1 ## 0 10 0 0 ## TTAGTCTGTATGAGCG-1 TTAGTCTGTCGATTTG-1 TTATTGCCACATATGC-1 TTATTGCTCAACTGAC-1 ## 2 1 2 8 ## TTATTGCTCATTTCGT-1 TTCAATCAGTGAGTGC-1 TTCAATCCATTAGGCT-1 TTCACCGAGGCTCAAG-1 ## 0 1 4 2 ## TTCACCGGTTCAACGT-1 TTCACGCGTCCGTACG-1 TTCAGGAAGAGAACCC-1 TTCAGGAAGGGCAGAG-1 ## 5 9 2 0 ## TTCAGGATCATCAGTG-1 TTCAGGATCATTTCCA-1 TTCATGTAGGACTAAT-1 TTCATTGGTCTCAGAT-1 ## 1 1 4 7 ## TTCCAATAGTCTGCGC-1 TTCCAATCATCAGCAT-1 TTCCACGGTAGGTGCA-1 TTCCACGGTATCGCTA-1 ## 9 5 0 2 ## TTCCGGTAGCTTAAGA-1 TTCCGGTCATCATCTT-1 TTCCGGTCATCCTAAG-1 TTCCGGTGTTGGTAGG-1 ## 4 7 6 0 ## TTCCGTGGTATGAAGT-1 TTCCTAAAGGCTCCCA-1 TTCCTCTAGGTCACAG-1 TTCCTCTGTACGTGTT-1 ## 3 7 0 6 ## TTCCTTCTCCACAAGT-1 TTCGATTTCTCCCATG-1 TTCGGTCTCGTAGTGT-1 TTCTAACCAAGCAGGT-1 ## 10 3 9 0 ## TTCTAACTCAAGCTTG-1 TTCTAGTAGCTAGAGC-1 TTCTAGTAGTTGGAAT-1 TTCTAGTGTTCTTCAT-1 ## 4 6 4 3 ## TTCTCTCAGCTGTTCA-1 TTCTGTAAGCATGTTC-1 TTCTGTAAGGAACATT-1 TTCTTCCAGTTCCATG-1 ## 9 1 2 0 ## TTCTTCCGTGCATTTG-1 TTCTTCCGTTAACAGA-1 TTCTTGAAGAAACCCG-1 TTGAACGAGAAGCCTG-1 ## 0 1 9 0 ## TTGAACGAGGGAGATA-1 TTGAACGTCCGATAAC-1 TTGACCCGTCGGTACC-1 TTGAGTGGTATATGGA-1 ## 0 1 3 0 ## TTGCATTCACATATGC-1 TTGCATTCAGGCTACC-1 TTGCATTGTTCCTAGA-1 TTGCATTTCACCTGTC-1 ## 1 1 2 3 ## TTGCCTGAGGTTAAAC-1 TTGCCTGCATTCACCC-1 TTGCGTCAGCATGCAG-1 TTGCGTCGTTCGGTCG-1 ## 0 1 1 0 ## TTGGATGTCTGGCCAG-1 TTGGGATGTACTGAGG-1 TTGGGATGTTTGTTCT-1 TTGGGCGAGGCCACTC-1 ## 3 2 8 9 ## TTGGGCGGTCCTTTGC-1 TTGGGCGGTTACAGCT-1 TTGGGTATCTTCCCAG-1 TTGGTTTCAGGTCAGA-1 ## 1 11 2 2 ## TTGTGTTCAATGGGTG-1 TTGTGTTGTCGAGCAA-1 TTGTTCAAGCAATTAG-1 TTGTTCACACCCTCTA-1 ## 4 0 1 6 ## TTGTTTGTCCTTCAGC-1 TTTACCACATCATCTT-1 TTTACTGTCAGGAAAT-1 TTTAGTCCAACGTATC-1 ## 1 6 4 1 ## TTTAGTCCACAGTCAT-1 TTTATGCAGCGCATCC-1 TTTATGCAGGATCATA-1 TTTATGCCAACAGAGC-1 ## 2 11 1 6 ## TTTATGCTCGTTCTGC-1 TTTCACAGTGACAGGT-1 TTTCACAGTGGCCTCA-1 TTTCAGTAGTCTACCA-1 ## 0 7 4 9 ## TTTCAGTCAGCTCGGT-1 TTTCATGAGGTTGACG-1 TTTCATGCATTGAAAG-1 TTTCATGTCTTTCCAA-1 ## 1 9 0 8 ## TTTCCTCCACCTGAAT-1 TTTCGATAGATAGCTA-1 TTTGACTAGACCCGCT-1 TTTGATCAGGAACGTC-1 ## 8 5 4 7 ## TTTGATCTCATTCTTG-1 TTTGGAGTCCACGTCT-1 TTTGGAGTCCGTAGGC-1 TTTGTTGGTCTAACTG-1 ## 8 0 4 5 ## Levels: 0 1 2 3 4 5 6 7 8 9 10 11 ``` --- ## Clustering We then visualise the clustering using the Dimplot fucntion which will use the active.ident slot by default. ``` r DimPlot(Nd1T.obj.filt, reduction = "umap") ``` <!-- --> --- # Compare clustering We can quickly compare the clusters from Bioconductor and Seurat. ``` r bioc.clusters <- data.frame(Bioc = sce.NeuroD1_filtered_QCed$label, row.names = sce.NeuroD1_filtered_QCed$Barcode) seurat.clusters <- as.data.frame(Nd1T.obj.filt@active.ident) colnames(seurat.clusters) <- "Seurat" clustersAll <- merge(bioc.clusters, seurat.clusters, by = 0, all = FALSE) ``` --- # Compare clustering ``` r tab <- table(Bioc = clustersAll$Bioc, Seurat = clustersAll$Seurat) rownames(tab) <- paste("Bioc", rownames(tab)) colnames(tab) <- paste("Seurat", colnames(tab)) library(pheatmap) pheatmap(log10(tab + 10), color = viridis::viridis(100), cluster_cols = FALSE, cluster_rows = FALSE) ``` <!-- --> --- # Find Markers The *FindAllMarkers* function allows us to identify markers across all clusters. ``` r Nd1T.markers <- FindAllMarkers(Nd1T.obj.filt, only.pos = TRUE) ``` ``` ## Calculating cluster 0 ``` ``` ## For a (much!) faster implementation of the Wilcoxon Rank Sum Test, ## (default method for FindMarkers) please install the presto package ## -------------------------------------------- ## install.packages('devtools') ## devtools::install_github('immunogenomics/presto') ## -------------------------------------------- ## After installation of presto, Seurat will automatically use the more ## efficient implementation (no further action necessary). ## This message will be shown once per session ``` ``` ## Calculating cluster 1 ``` ``` ## Calculating cluster 2 ``` ``` ## Calculating cluster 3 ``` ``` ## Calculating cluster 4 ``` ``` ## Calculating cluster 5 ``` ``` ## Calculating cluster 6 ``` ``` ## Calculating cluster 7 ``` ``` ## Calculating cluster 8 ``` ``` ## Calculating cluster 9 ``` ``` ## Calculating cluster 10 ``` ``` ## Calculating cluster 11 ``` ``` r Nd1T.markers[Nd1T.markers$cluster == 1, ] ``` ``` ## p_val avg_log2FC pct.1 pct.2 p_val_adj cluster ## Pzp 2.699922e-270 5.6606543 0.976 0.055 1.077404e-265 1 ## Habp2 1.065667e-230 4.3811570 0.935 0.074 4.252544e-226 1 ## Crp 1.349748e-226 4.2773148 0.947 0.080 5.386168e-222 1 ## Adamts5 9.886811e-223 4.6271800 0.788 0.027 3.945332e-218 1 ## Cyp2j5 3.468656e-222 4.5128990 0.829 0.040 1.384167e-217 1 ## Ugt8a 5.906877e-190 4.1730113 0.665 0.020 2.357139e-185 1 ## Basp1 6.426782e-184 4.3649501 0.665 0.022 2.564607e-179 1 ## Mc4r 7.313401e-184 4.9362268 0.657 0.021 2.918413e-179 1 ## Col17a1 6.137550e-182 4.8601391 0.645 0.019 2.449189e-177 1 ## Kcnk16 3.744829e-175 3.1939737 0.927 0.115 1.494374e-170 1 ## Galnt13 1.191308e-164 3.1462945 0.878 0.103 4.753915e-160 1 ## Tlr2 1.506014e-162 3.8371137 0.694 0.048 6.009747e-158 1 ## Casr 1.028930e-160 2.9800779 0.927 0.143 4.105946e-156 1 ## Rbm24 2.789006e-160 4.3233404 0.608 0.024 1.112953e-155 1 ## Gm33649 5.992132e-156 4.1462566 0.547 0.014 2.391160e-151 1 ## Onecut3 6.223527e-154 2.8948211 0.984 0.205 2.483498e-149 1 ## Crtac1 9.804624e-146 5.4900467 0.478 0.007 3.912535e-141 1 ## Tlr5 1.882859e-142 4.3721675 0.510 0.014 7.513550e-138 1 ## Cltrn 5.535703e-142 2.7783686 0.935 0.189 2.209022e-137 1 ## Smbd1 7.888393e-141 3.5124344 0.739 0.087 3.147863e-136 1 ## Zfp618 2.533112e-136 2.8852924 0.865 0.150 1.010839e-131 1 ## Pgc 4.878338e-133 3.5945738 0.829 0.136 1.946701e-128 1 ## E030025P04Rik 1.594135e-129 3.9813785 0.482 0.015 6.361396e-125 1 ## Cyp2j6 1.181605e-127 3.1630302 0.988 0.435 4.715193e-123 1 ## Tril 2.336380e-123 3.2223425 0.714 0.089 9.323324e-119 1 ## Agr2 1.023802e-121 3.1816234 1.000 0.572 4.085481e-117 1 ## Gm53467 8.818278e-119 2.4504236 0.939 0.253 3.518934e-114 1 ## Agr31 1.358279e-118 2.3179350 1.000 0.492 5.420211e-114 1 ## D930028M14Rik 4.800545e-118 2.4720201 0.947 0.256 1.915658e-113 1 ## Slc39a8 9.115504e-118 2.6002583 0.927 0.235 3.637542e-113 1 ## Cck 8.818541e-117 3.3514533 1.000 0.989 3.519039e-112 1 ## Scg2 1.040437e-115 2.5621438 1.000 0.611 4.151862e-111 1 ## Scn9a 3.758629e-114 5.3441851 0.371 0.004 1.499881e-109 1 ## Gm12511 6.827378e-114 2.7522393 0.771 0.128 2.724465e-109 1 ## Etv1 8.327534e-114 2.1706393 0.967 0.258 3.323103e-109 1 ## Upp1 2.143444e-113 2.2047997 0.996 0.665 8.553415e-109 1 ## Rflna 2.836208e-113 2.5034590 0.918 0.231 1.131789e-108 1 ## Parm1 3.147554e-113 2.2340147 0.976 0.301 1.256031e-108 1 ## Scn3b 6.169439e-112 3.0083149 0.596 0.058 2.461915e-107 1 ## U90926 5.214799e-109 4.1650117 0.494 0.033 2.080966e-104 1 ## Cpn1 6.472940e-108 2.3458756 0.931 0.284 2.583027e-103 1 ## Cux2 9.328695e-107 3.7957808 0.429 0.018 3.722616e-102 1 ## Cst3 1.001014e-104 1.6994862 1.000 0.783 3.994545e-100 1 ## Tle1 6.974413e-104 2.3750018 0.967 0.449 2.783140e-99 1 ## Fmo5 1.919841e-103 2.1599524 0.988 0.450 7.661125e-99 1 ## 3110004A20Rik 3.831722e-103 4.4818634 0.396 0.015 1.529049e-98 1 ## Tmem176a 4.401922e-103 1.8917325 0.996 0.595 1.756587e-98 1 ## Gm11992 3.714380e-101 2.0167454 0.927 0.229 1.482223e-96 1 ## Proc 6.425485e-101 3.7477873 0.433 0.024 2.564090e-96 1 ## Egfl6 6.839051e-101 4.5368789 0.371 0.011 2.729123e-96 1 ## Spart 9.053020e-101 2.4797895 0.861 0.229 3.612608e-96 1 ## Vkorc1 1.172423e-99 2.4020757 0.910 0.299 4.678554e-95 1 ## Arx 1.658712e-99 2.1595432 0.927 0.249 6.619089e-95 1 ## Rdh7 7.457938e-99 2.3733200 0.669 0.103 2.976090e-94 1 ## Lyzl4 8.956323e-99 2.5001093 0.592 0.068 3.574021e-94 1 ## Tec 1.597456e-97 2.4654281 0.861 0.238 6.374647e-93 1 ## Tff31 6.100035e-96 1.8674942 0.996 0.881 2.434219e-91 1 ## Slc41a2 1.583432e-95 2.1924161 0.882 0.269 6.318686e-91 1 ## Dclk3 1.776045e-95 2.3645957 0.710 0.126 7.087306e-91 1 ## S100a11 8.175745e-93 2.0034438 0.980 0.405 3.262531e-88 1 ## Cit 2.648367e-92 1.9336515 0.833 0.204 1.056831e-87 1 ## Rfxap 3.342280e-92 2.5108874 0.882 0.323 1.333737e-87 1 ## Cav2 7.141018e-90 2.5133650 0.620 0.090 2.849623e-85 1 ## Aldoc 8.099228e-89 2.3599038 0.796 0.207 3.231997e-84 1 ## Ms4a10 1.049211e-88 1.9536551 0.882 0.280 4.186878e-84 1 ## Homer21 1.063782e-87 1.8745146 0.963 0.423 4.245021e-83 1 ## Fgl2 4.695166e-87 2.1447225 0.931 0.392 1.873606e-82 1 ## Slc8a1 5.347902e-87 1.7045991 0.996 0.549 2.134080e-82 1 ## Pla2g12a 1.823729e-86 1.8501690 0.980 0.470 7.277589e-82 1 ## Fars2 3.438234e-85 2.2589860 0.914 0.359 1.372027e-80 1 ## Rbp4 5.215000e-85 1.6486007 1.000 0.495 2.081046e-80 1 ## Igfbp7 2.674851e-84 2.7932609 0.482 0.052 1.067399e-79 1 ## Oxr1 1.321102e-83 1.7049341 0.996 0.691 5.271856e-79 1 ## Itm2b 4.032685e-82 1.2579368 1.000 0.877 1.609243e-77 1 ## Slc25a4 2.835038e-81 1.6469967 0.976 0.617 1.131322e-76 1 ## Tm4sf41 2.335096e-80 1.1351982 1.000 0.918 9.318199e-76 1 ## Ms4a8a1 1.224548e-79 1.5883277 0.992 0.695 4.886558e-75 1 ## Dapp1 1.459266e-79 1.6810650 0.886 0.287 5.823200e-75 1 ## Nenf 2.167489e-79 1.4540964 0.992 0.680 8.649367e-75 1 ## Aig1 2.255183e-79 1.5528806 0.992 0.595 8.999308e-75 1 ## Ly6e 3.008964e-79 2.4690674 0.824 0.242 1.200727e-74 1 ## Prox1 8.300380e-79 1.6788435 0.963 0.403 3.312267e-74 1 ## Ifitm2 1.086416e-78 1.6297997 0.906 0.314 4.335342e-74 1 ## Bace2 1.732120e-78 1.4699255 0.898 0.247 6.912026e-74 1 ## Marveld1 3.330427e-78 3.6512724 0.376 0.027 1.329007e-73 1 ## Grpr 4.003577e-78 1.6315261 0.571 0.086 1.597627e-73 1 ## Me3 1.688502e-77 2.6871858 0.376 0.027 6.737968e-73 1 ## Gm27162 2.463489e-77 1.8021242 0.763 0.177 9.830553e-73 1 ## Tmem176b 2.662862e-77 1.3494808 1.000 0.716 1.062615e-72 1 ## Trpm3 8.390992e-77 5.0879826 0.261 0.004 3.348425e-72 1 ## Cyp2j9 1.065010e-75 3.1894100 0.433 0.046 4.249923e-71 1 ## Prss23 4.723288e-75 3.0474488 0.490 0.070 1.884828e-70 1 ## Lgals21 5.046157e-75 1.1905090 0.996 0.950 2.013669e-70 1 ## Qpct 6.200043e-75 1.7982531 0.980 0.478 2.474127e-70 1 ## Gm46558 1.413407e-74 5.0383671 0.241 0.002 5.640203e-70 1 ## Sting1 1.566197e-74 3.6273476 0.339 0.022 6.249911e-70 1 ## Ugt2b34 8.978780e-74 1.4485356 0.976 0.520 3.582982e-69 1 ## Myl7 1.000315e-72 1.6135431 0.996 0.786 3.991759e-68 1 ## Dner 3.805951e-72 1.7775049 0.910 0.328 1.518765e-67 1 ## Tspan12 5.812781e-72 1.7983967 0.906 0.347 2.319590e-67 1 ## Casp6 8.859097e-71 1.6171429 0.976 0.579 3.535223e-66 1 ## Plac81 1.478954e-70 1.0396146 1.000 0.949 5.901765e-66 1 ## Klkb11 2.267775e-70 2.5294420 0.535 0.090 9.049556e-66 1 ## Gm46905 5.162225e-70 4.0048248 0.245 0.005 2.059986e-65 1 ## Serpina1b1 2.397324e-69 1.4873746 0.861 0.287 9.566520e-65 1 ## Wars1 5.810624e-69 1.7504428 0.837 0.273 2.318729e-64 1 ## Tshz2 6.130726e-68 1.9558450 0.567 0.102 2.446466e-63 1 ## Abca8b 2.200108e-67 3.9648622 0.261 0.009 8.779532e-63 1 ## Krt201 1.404465e-66 1.3187618 0.996 0.767 5.604519e-62 1 ## Atp8a2 2.722469e-66 2.1714690 0.682 0.181 1.086401e-61 1 ## Sstr2 3.489732e-65 2.2573114 0.478 0.072 1.392578e-60 1 ## Gm17619 5.929589e-65 2.9736915 0.400 0.050 2.366203e-60 1 ## Axin2 6.203368e-65 1.5413060 0.894 0.346 2.475454e-60 1 ## Tmed6 6.574956e-65 2.1321333 0.727 0.230 2.623736e-60 1 ## Tpst2 7.305380e-65 1.5909708 0.931 0.437 2.915212e-60 1 ## Serpine2 7.334110e-65 3.5879984 0.282 0.015 2.926677e-60 1 ## Anxa13 7.712752e-65 1.3364224 0.906 0.345 3.077774e-60 1 ## Apom 1.340645e-64 2.1845451 0.596 0.130 5.349843e-60 1 ## Dio1 1.390741e-64 1.8076877 0.784 0.247 5.549751e-60 1 ## Upk1b 3.094013e-64 2.9220043 0.445 0.066 1.234666e-59 1 ## Sct 3.519956e-64 1.3126059 1.000 0.995 1.404638e-59 1 ## Egfl7 3.531622e-64 2.3241940 0.506 0.087 1.409294e-59 1 ## Psap 1.080772e-63 1.2022266 0.996 0.760 4.312822e-59 1 ## Gclm 2.227809e-63 1.1377615 0.992 0.766 8.890072e-59 1 ## Gm52807 2.562262e-63 4.7797797 0.216 0.003 1.022471e-58 1 ## Smarca1 2.724802e-63 1.6239375 0.698 0.178 1.087332e-58 1 ## Cryba4 3.109212e-63 2.9562990 0.331 0.028 1.240731e-58 1 ## Rflnb 5.236190e-63 1.9436153 0.604 0.136 2.089502e-58 1 ## Shf 1.191860e-62 1.3455651 0.865 0.309 4.756116e-58 1 ## Gast 2.102266e-62 7.5692332 0.514 0.101 8.389091e-58 1 ## Bsg1 2.404722e-62 0.7960773 0.996 0.899 9.596043e-58 1 ## Ano6 2.841087e-62 1.2576955 0.939 0.424 1.133736e-57 1 ## Spry1 4.864743e-62 2.4138687 0.457 0.073 1.941276e-57 1 ## F51 2.099135e-61 1.6080218 0.829 0.277 8.376597e-57 1 ## Ndrg11 2.241179e-61 1.2343521 0.988 0.745 8.943424e-57 1 ## Aga1 2.319167e-61 1.7052487 0.918 0.488 9.254636e-57 1 ## Lrpap1 3.616715e-61 1.7566745 0.865 0.385 1.443250e-56 1 ## Trf1 2.138803e-60 1.7802646 0.890 0.357 8.534892e-56 1 ## Tmed3 4.572586e-60 1.3059738 0.996 0.609 1.824690e-55 1 ## Uts2b 5.282028e-60 2.7514124 0.327 0.029 2.107793e-55 1 ## Kcnj6 9.580984e-60 1.4296888 0.686 0.173 3.823292e-55 1 ## Cops6 1.728726e-59 1.5729054 0.947 0.488 6.898480e-55 1 ## Cpm 2.350687e-59 1.8965689 0.767 0.270 9.380418e-55 1 ## Cpe 8.610885e-59 1.1755585 0.996 0.905 3.436173e-54 1 ## Ssr4 2.300570e-58 1.1378480 0.988 0.747 9.180426e-54 1 ## Rps20 3.118018e-58 0.5205074 1.000 0.892 1.244245e-53 1 ## Col6a5 3.196382e-58 1.5415562 0.404 0.054 1.275516e-53 1 ## Tmem254 3.358666e-58 1.4494973 0.914 0.418 1.340276e-53 1 ## Gnai1 5.648399e-58 1.7128553 0.694 0.184 2.253994e-53 1 ## Atoh8 7.000848e-58 3.5094188 0.306 0.027 2.793689e-53 1 ## Rps24 7.324649e-58 0.6755523 1.000 0.891 2.922901e-53 1 ## Txndc5 3.138794e-57 1.3086817 0.967 0.587 1.252536e-52 1 ## Cyp2c55 2.650528e-56 1.8353372 0.547 0.121 1.057693e-51 1 ## Gm53234 4.294674e-56 3.4645442 0.253 0.015 1.713790e-51 1 ## Smad9 9.065755e-56 1.8116976 0.608 0.146 3.617689e-51 1 ## Rpl32 9.323478e-56 0.5952376 1.000 0.869 3.720534e-51 1 ## Ppic1 9.466628e-56 1.6639835 0.824 0.306 3.777658e-51 1 ## Bambi 1.402100e-55 1.2894428 0.857 0.299 5.595081e-51 1 ## Rasgrf2 1.541429e-55 1.3134649 0.853 0.319 6.151071e-51 1 ## 4732490B19Rik 1.864822e-55 2.3092791 0.347 0.041 7.441572e-51 1 ## Gm9767 2.451550e-55 2.6488372 0.253 0.015 9.782910e-51 1 ## Mtcl2 1.059244e-54 2.0098361 0.502 0.099 4.226915e-50 1 ## Ihh 2.983828e-54 1.8015607 0.653 0.190 1.190696e-49 1 ## Rps27a 5.738969e-54 0.6680022 1.000 0.855 2.290136e-49 1 ## Rps18 7.330043e-54 0.5485342 1.000 0.862 2.925054e-49 1 ## Pcbd11 1.493304e-53 1.1670292 0.976 0.667 5.959029e-49 1 ## Pabpc1l 1.795249e-53 2.3910452 0.453 0.083 7.163942e-49 1 ## Exoc7 2.411724e-53 2.0404750 0.776 0.345 9.623986e-49 1 ## Gcnt3 2.482683e-53 1.2215079 0.878 0.338 9.907146e-49 1 ## Hunk1 3.764435e-52 1.3687881 0.849 0.335 1.502198e-47 1 ## Cited2 3.157411e-51 1.1775566 0.984 0.684 1.259965e-46 1 ## Gnas 4.731274e-51 0.6356772 1.000 0.914 1.888015e-46 1 ## Fxyd31 8.274089e-51 0.9435968 1.000 0.823 3.301775e-46 1 ## Gimd11 9.583149e-51 1.4135039 0.894 0.427 3.824156e-46 1 ## Pxylp1 9.976340e-51 2.2974431 0.461 0.093 3.981058e-46 1 ## Gm30524 1.211363e-50 1.8637308 0.392 0.059 4.833944e-46 1 ## 4930573H18Rik 2.106652e-50 3.4027268 0.220 0.012 8.406594e-46 1 ## Mill2 6.839570e-50 1.9613373 0.441 0.083 2.729330e-45 1 ## Abcb1a 2.546123e-49 1.0566381 0.967 0.600 1.016030e-44 1 ## Gfra1 3.078817e-49 1.8714073 0.457 0.089 1.228602e-44 1 ## Gm2102 5.364457e-49 4.5598845 0.163 0.002 2.140687e-44 1 ## Anxa2 6.714237e-49 0.9336810 0.988 0.603 2.679316e-44 1 ## Rpl38 7.057253e-49 0.6037217 1.000 0.905 2.816197e-44 1 ## Fut2 4.612345e-48 1.3789870 0.637 0.174 1.840556e-43 1 ## Apcs 1.212976e-47 5.1409002 0.163 0.003 4.840380e-43 1 ## Ghrl 1.788400e-47 1.0158195 0.939 0.758 7.136609e-43 1 ## Cfi 1.802052e-47 1.9864944 0.608 0.184 7.191088e-43 1 ## Cldn16 1.836310e-47 5.1617056 0.171 0.004 7.327794e-43 1 ## Sis1 1.916725e-47 0.9440739 0.996 0.855 7.648692e-43 1 ## Pde3a 2.040256e-47 1.3549620 0.653 0.202 8.141641e-43 1 ## Efcab151 3.031130e-47 1.6047977 0.804 0.335 1.209572e-42 1 ## C2 5.145329e-47 1.8115322 0.506 0.119 2.053244e-42 1 ## Camk4 6.434404e-47 4.6964399 0.188 0.008 2.567649e-42 1 ## Plgrkt 9.153068e-47 1.2240517 0.918 0.477 3.652532e-42 1 ## Gm30986 1.076640e-46 3.1529339 0.220 0.015 4.296333e-42 1 ## Cyp4f14 1.113060e-46 0.5913869 0.833 0.334 4.441664e-42 1 ## Nfix 1.136348e-46 1.5569512 0.645 0.194 4.534598e-42 1 ## Spint21 1.239720e-46 0.6833710 1.000 0.892 4.947103e-42 1 ## Nefl 2.041210e-46 2.0595865 0.392 0.071 8.145448e-42 1 ## Trappc9 2.362913e-46 1.7007704 0.718 0.277 9.429203e-42 1 ## Sema4g 2.990686e-46 1.0452624 0.963 0.569 1.193433e-41 1 ## Pnma8b 3.326133e-46 1.9138907 0.494 0.117 1.327294e-41 1 ## Cdkl1 6.683605e-46 2.1134866 0.445 0.094 2.667093e-41 1 ## 2610524H06Rik 1.402422e-45 1.5091068 0.755 0.282 5.596365e-41 1 ## Slc12a8 1.503757e-45 1.4864049 0.739 0.273 6.000743e-41 1 ## Nbdy 2.239100e-45 1.3032475 0.829 0.365 8.935129e-41 1 ## Rpl19 2.311746e-45 0.5990009 1.000 0.904 9.225023e-41 1 ## Atp5mk1 2.872883e-45 0.8281701 0.996 0.796 1.146424e-40 1 ## H2-D1 4.304026e-45 0.6642803 1.000 0.891 1.717521e-40 1 ## Rtn4r 1.111387e-44 1.5305343 0.469 0.103 4.434989e-40 1 ## Gabarapl21 1.374604e-44 0.8722304 1.000 0.760 5.485359e-40 1 ## Rps3 2.361735e-44 0.4684751 1.000 0.849 9.424504e-40 1 ## Pfkfb3 3.064788e-44 2.1078254 0.498 0.130 1.223004e-39 1 ## Tspo 3.893485e-44 1.0826240 0.947 0.535 1.553695e-39 1 ## Rps10 4.746915e-44 0.4827791 1.000 0.879 1.894256e-39 1 ## Snhg18 6.834945e-44 1.4010768 0.812 0.375 2.727485e-39 1 ## Ebpl 7.344838e-44 1.8199124 0.616 0.203 2.930957e-39 1 ## Nostrin1 1.207339e-43 0.9931323 0.980 0.700 4.817888e-39 1 ## Pir 1.411112e-43 2.7107409 0.331 0.053 5.631042e-39 1 ## Lgals3bp 1.768105e-43 0.7729091 0.996 0.868 7.055622e-39 1 ## Tmod2 1.954850e-43 1.3460208 0.641 0.194 7.800829e-39 1 ## Tpst11 2.398998e-43 1.1531554 0.931 0.470 9.573202e-39 1 ## Rps2 3.740603e-43 0.2769781 0.996 0.835 1.492687e-38 1 ## Rpl39 4.525499e-43 0.4400616 1.000 0.826 1.805900e-38 1 ## Olfm11 4.972244e-43 1.0140321 0.967 0.552 1.984174e-38 1 ## Cr1l 4.994419e-43 1.2622647 0.820 0.396 1.993023e-38 1 ## Fads2 6.308745e-43 1.2529416 0.796 0.320 2.517505e-38 1 ## Pcbd2 6.349248e-43 1.2818686 0.841 0.389 2.533667e-38 1 ## Efcc1 6.366714e-43 1.6186749 0.486 0.118 2.540637e-38 1 ## Krt79 7.809877e-43 6.7616835 0.139 0.001 3.116532e-38 1 ## Ldb3 1.292229e-42 1.5909123 0.567 0.172 5.156639e-38 1 ## Gm39858 1.810936e-42 1.7818897 0.506 0.127 7.226540e-38 1 ## Rpl12 1.895198e-42 0.4276460 0.992 0.752 7.562786e-38 1 ## Cyp2c681 3.210891e-42 1.3589325 0.943 0.577 1.281306e-37 1 ## Col11a2 3.970256e-42 1.9756894 0.331 0.053 1.584331e-37 1 ## Hnf1aos1 4.091696e-42 1.3839968 0.678 0.237 1.632791e-37 1 ## Cldn1 5.146380e-42 2.9276384 0.278 0.036 2.053663e-37 1 ## Slit1 5.634545e-42 1.9720141 0.273 0.034 2.248465e-37 1 ## Mycn 6.116033e-42 3.5164143 0.224 0.020 2.440603e-37 1 ## Ccdc85b 6.246715e-42 1.3282818 0.788 0.336 2.492752e-37 1 ## Tm7sf2 1.475556e-41 1.6762902 0.535 0.149 5.888208e-37 1 ## Pepd 2.022208e-41 0.5576880 0.951 0.568 8.069622e-37 1 ## 5033421B08Rik 3.029663e-41 1.5360792 0.510 0.139 1.208987e-36 1 ## Maged2 3.241248e-41 1.1092219 0.702 0.261 1.293420e-36 1 ## Mgll1 3.301249e-41 0.6921027 0.971 0.478 1.317364e-36 1 ## Cnpy2 4.646311e-41 0.9769550 0.959 0.602 1.854110e-36 1 ## Slc16a10 4.762245e-41 1.0985370 0.898 0.435 1.900374e-36 1 ## Bhlhe41 5.653975e-41 3.6861646 0.180 0.010 2.256219e-36 1 ## Lysmd21 5.833553e-41 1.3750668 0.743 0.293 2.327879e-36 1 ## Zfp395 8.267744e-41 1.8122712 0.486 0.132 3.299243e-36 1 ## Pglyrp1 9.387021e-41 1.2635429 0.767 0.310 3.745891e-36 1 ## Amotl2 1.017190e-40 1.9665896 0.424 0.094 4.059096e-36 1 ## Hgfac 1.068820e-40 1.1588171 0.739 0.284 4.265128e-36 1 ## Endod1 1.793231e-40 1.9221853 0.408 0.093 7.155889e-36 1 ## B3gnt9 2.170193e-40 3.1320530 0.216 0.019 8.660153e-36 1 ## Ctdspl 2.306939e-40 1.0131998 0.959 0.627 9.205842e-36 1 ## Elapor11 3.106513e-40 0.7005788 0.992 0.823 1.239654e-35 1 ## Sh3pxd2a 3.416424e-40 1.4835701 0.518 0.138 1.363324e-35 1 ## Rpl15 3.787369e-40 0.5448505 1.000 0.823 1.511350e-35 1 ## Ywhah 6.551044e-40 0.8801902 0.992 0.739 2.614194e-35 1 ## Nog 6.787547e-40 5.3783536 0.139 0.003 2.708571e-35 1 ## Rpl37a1 6.990589e-40 0.6747938 1.000 0.911 2.789595e-35 1 ## Tpmt 8.034393e-40 1.4862230 0.645 0.228 3.206125e-35 1 ## Ttr1 9.974639e-40 0.9020619 0.984 0.639 3.980380e-35 1 ## Cap2 1.363737e-39 3.1851436 0.216 0.020 5.441992e-35 1 ## Calml41 1.872097e-39 0.7380436 0.984 0.822 7.470602e-35 1 ## Rps7 1.902562e-39 0.4040669 1.000 0.864 7.592174e-35 1 ## Uchl1 1.928426e-39 2.8280554 0.261 0.034 7.695385e-35 1 ## Tdrd12 2.088979e-39 1.7874130 0.351 0.064 8.336070e-35 1 ## Arsb1 2.183267e-39 1.5024028 0.649 0.229 8.712326e-35 1 ## H2-K1 2.689523e-39 0.5645869 1.000 0.929 1.073254e-34 1 ## Hexb 4.191163e-39 0.9790959 0.971 0.622 1.672484e-34 1 ## Fam222a 4.416271e-39 1.5585381 0.649 0.237 1.762313e-34 1 ## Ppp1r14a 7.748390e-39 4.0067024 0.155 0.006 3.091995e-34 1 ## Snx18 8.950700e-39 1.1453971 0.829 0.375 3.571777e-34 1 ## Ypel3 1.024671e-38 1.0282424 0.922 0.530 4.088948e-34 1 ## Qsox1 1.370082e-38 1.0289799 0.694 0.251 5.467310e-34 1 ## Cpq 1.413468e-38 1.0916543 0.927 0.574 5.640444e-34 1 ## Ptp4a3 2.304598e-38 1.3662251 0.571 0.180 9.196499e-34 1 ## Fam174a1 2.737505e-38 1.2755907 0.792 0.388 1.092401e-33 1 ## Muc4 3.629919e-38 0.9382476 0.951 0.589 1.448519e-33 1 ## Prdx11 4.674057e-38 0.6948415 1.000 0.857 1.865182e-33 1 ## Gm30648 5.263842e-38 1.0460127 0.755 0.290 2.100536e-33 1 ## Rpl37 5.940662e-38 0.5168139 0.992 0.845 2.370621e-33 1 ## Rpl27a 5.971242e-38 0.3494292 1.000 0.846 2.382824e-33 1 ## Nkx6-3 6.038541e-38 2.9154675 0.265 0.038 2.409680e-33 1 ## Klf7 8.370308e-38 0.9054215 0.947 0.574 3.340171e-33 1 ## Epha4 1.218606e-37 3.1115231 0.245 0.031 4.862846e-33 1 ## Kcnj3 1.278056e-37 0.9065844 0.706 0.243 5.100082e-33 1 ## Ctsf 1.628573e-37 1.4095009 0.649 0.229 6.498821e-33 1 ## Ggact 1.688269e-37 1.3866874 0.678 0.247 6.737037e-33 1 ## Ces2a 2.078520e-37 1.1295049 0.661 0.252 8.294335e-33 1 ## Mrpl15 3.150073e-37 1.0372414 0.951 0.563 1.257036e-32 1 ## Vsig2 3.164576e-37 1.8966708 0.588 0.214 1.262824e-32 1 ## Rnf130 3.188723e-37 1.0594787 0.841 0.358 1.272460e-32 1 ## Blnk 4.507642e-37 1.1970214 0.653 0.239 1.798774e-32 1 ## Riiad11 4.703027e-37 1.0823788 0.861 0.410 1.876743e-32 1 ## Acaa2 5.109312e-37 0.8198237 0.894 0.462 2.038871e-32 1 ## Tomm7 5.231606e-37 0.7414403 0.980 0.715 2.087672e-32 1 ## Atp5mc2 6.645033e-37 0.6572135 0.992 0.751 2.651700e-32 1 ## Svip1 8.547722e-37 1.0055335 0.971 0.660 3.410968e-32 1 ## Fxyd5 8.925146e-37 2.2850444 0.261 0.036 3.561580e-32 1 ## 9330162012Rik 9.347575e-37 1.6088384 0.461 0.126 3.730150e-32 1 ## Rpl13 1.109664e-36 0.2984148 0.996 0.876 4.428115e-32 1 ## Rpl3 1.151154e-36 0.4702908 1.000 0.804 4.593680e-32 1 ## Rps4x 1.857093e-36 0.3060136 1.000 0.843 7.410731e-32 1 ## Hagh 1.899564e-36 0.9359687 0.865 0.427 7.580210e-32 1 ## Rap1gap 2.030925e-36 0.8601867 0.767 0.289 8.104406e-32 1 ## Mpc2 2.948566e-36 0.8495130 0.959 0.623 1.176625e-31 1 ## Usp25 3.861924e-36 1.1312970 0.763 0.351 1.541101e-31 1 ## Klf12 4.038152e-36 0.8968232 0.829 0.389 1.611425e-31 1 ## Tspan11 4.295184e-36 0.7719039 0.992 0.745 1.713993e-31 1 ## Commd1 4.326315e-36 0.9716163 0.910 0.523 1.726416e-31 1 ## Cped1 4.866829e-36 1.2328366 0.429 0.103 1.942108e-31 1 ## Ifi271 5.946767e-36 0.9291664 0.967 0.635 2.373057e-31 1 ## Cfap69 6.686697e-36 2.2975039 0.245 0.032 2.668326e-31 1 ## Tulp2 9.336578e-36 1.8708780 0.465 0.133 3.725761e-31 1 ## Bcl11a 9.522313e-36 1.2569785 0.661 0.246 3.799879e-31 1 ## Kif2a 1.320346e-35 1.0911610 0.873 0.492 5.268842e-31 1 ## Trp53inp1 1.402144e-35 1.0945251 0.857 0.422 5.595256e-31 1 ## Slc50a1 1.599153e-35 0.9798110 0.882 0.440 6.381420e-31 1 ## Mettl261 1.731153e-35 0.8450695 0.976 0.679 6.908167e-31 1 ## Pdzk1ip1 1.978514e-35 1.5991213 0.547 0.180 7.895262e-31 1 ## 4732419C18Rik 2.010764e-35 1.1711648 0.510 0.143 8.023955e-31 1 ## Rps3a1 2.050604e-35 0.4472840 1.000 0.913 8.182935e-31 1 ## Rpl23 2.275405e-35 0.3414371 1.000 0.883 9.080005e-31 1 ## Gm39460 2.681541e-35 3.2144401 0.151 0.008 1.070069e-30 1 ## Bcam1 4.553599e-35 0.7894596 0.951 0.500 1.817114e-30 1 ## Rps16 4.577928e-35 0.3428853 0.996 0.860 1.826822e-30 1 ## Rps15a 4.843569e-35 0.3682422 1.000 0.806 1.932826e-30 1 ## Mia 5.088233e-35 1.6619755 0.510 0.157 2.030459e-30 1 ## Atp5if1 7.750252e-35 0.5486023 1.000 0.891 3.092738e-30 1 ## Rnaset2a 1.137939e-34 1.4096674 0.563 0.197 4.540944e-30 1 ## Grcc101 1.563999e-34 0.8174604 0.980 0.706 6.241140e-30 1 ## Tcaf2 1.583057e-34 2.4801425 0.249 0.037 6.317189e-30 1 ## Tsc22d11 1.680242e-34 0.7669682 1.000 0.808 6.705005e-30 1 ## Urah 1.688806e-34 1.2175588 0.702 0.301 6.739181e-30 1 ## Shisal1 1.711109e-34 1.1913223 0.694 0.269 6.828181e-30 1 ## Msrb1 1.772809e-34 1.1690476 0.812 0.428 7.074394e-30 1 ## Marchf11 2.117545e-34 2.2506557 0.245 0.034 8.450063e-30 1 ## Mtcl1 2.888601e-34 1.2419787 0.506 0.152 1.152696e-29 1 ## Ppia1 3.827515e-34 0.5679656 0.996 0.861 1.527370e-29 1 ## Cga 4.457633e-34 6.2576762 0.102 0.000 1.778819e-29 1 ## Grb14 4.459405e-34 1.5189151 0.498 0.152 1.779526e-29 1 ## Rps13 5.526397e-34 0.5193778 1.000 0.813 2.205309e-29 1 ## Pla2g2d 7.054705e-34 1.2756022 0.420 0.108 2.815180e-29 1 ## Gcg 7.352591e-34 0.6440198 0.629 0.250 2.934051e-29 1 ## Scgn 8.128487e-34 0.4359863 0.922 0.342 3.243673e-29 1 ## Gde1 1.313310e-33 0.7507314 0.845 0.392 5.240765e-29 1 ## Gm41848 1.664858e-33 5.6101120 0.110 0.001 6.643614e-29 1 ## Gm142071 1.976618e-33 1.2807299 0.727 0.328 7.887693e-29 1 ## Rpl11 2.947544e-33 0.4628744 1.000 0.869 1.176217e-28 1 ## Slc22a15 3.463654e-33 1.6676405 0.310 0.061 1.382171e-28 1 ## Dip2a 3.572766e-33 1.4380247 0.424 0.117 1.425712e-28 1 ## Ikbip 3.728398e-33 1.9505285 0.380 0.093 1.487817e-28 1 ## Lsm6 4.016390e-33 0.7993416 0.943 0.600 1.602741e-28 1 ## Trim2 4.133048e-33 0.7170364 0.771 0.320 1.649293e-28 1 ## Romo11 5.636198e-33 0.6991394 0.996 0.741 2.249125e-28 1 ## Rpl17 5.651397e-33 0.3628479 1.000 0.834 2.255190e-28 1 ## Ceacam10 5.878007e-33 0.8301021 0.849 0.430 2.345619e-28 1 ## Glod51 6.283839e-33 0.8574353 0.935 0.537 2.507566e-28 1 ## Cdkl5 6.536730e-33 2.0530677 0.302 0.059 2.608482e-28 1 ## Agfg2 6.862877e-33 1.2146237 0.645 0.242 2.738631e-28 1 ## Tmem263 7.171557e-33 1.0043139 0.796 0.370 2.861810e-28 1 ## Lin7a 7.481318e-33 1.2154195 0.486 0.139 2.985420e-28 1 ## G430095P16Rik 8.558917e-33 4.0446741 0.139 0.007 3.415436e-28 1 ## Ret1 8.574065e-33 0.6949308 0.865 0.420 3.421481e-28 1 ## Eef1a1 9.666518e-33 0.4392696 1.000 0.955 3.857424e-28 1 ## Rac3 1.159170e-32 1.4367247 0.510 0.165 4.625670e-28 1 ## Nhlh1 1.265039e-32 2.1544075 0.265 0.043 5.048138e-28 1 ## Tbc1d9 1.580508e-32 1.2404569 0.641 0.238 6.307019e-28 1 ## Rpl34 1.870129e-32 0.4902920 1.000 0.807 7.462748e-28 1 ## Leprotl11 2.062896e-32 0.9723728 0.910 0.568 8.231987e-28 1 ## Sh3bgrl3 2.181509e-32 0.6913371 0.967 0.648 8.705311e-28 1 ## Gm53846 2.530536e-32 2.3848649 0.216 0.027 1.009811e-27 1 ## Lbp 2.767674e-32 3.2469131 0.155 0.011 1.104440e-27 1 ## Cd59a 2.850949e-32 1.0450188 0.657 0.259 1.137671e-27 1 ## Rnaset2b 2.857319e-32 1.5586598 0.502 0.168 1.140213e-27 1 ## Ero1b 3.256420e-32 1.2629842 0.522 0.171 1.299474e-27 1 ## P2rx4 4.810571e-32 1.3787344 0.588 0.220 1.919658e-27 1 ## Diras2 5.009548e-32 2.8785543 0.167 0.014 1.999060e-27 1 ## Sgpp2 5.468354e-32 0.9357284 0.910 0.528 2.182147e-27 1 ## F13b 5.519631e-32 2.4135657 0.118 0.003 2.202609e-27 1 ## Mgst2 5.893623e-32 0.6131699 0.645 0.240 2.351850e-27 1 ## Rpl35a 6.576440e-32 0.4303394 1.000 0.793 2.624328e-27 1 ## Uba52 9.501528e-32 0.3023864 1.000 0.938 3.791585e-27 1 ## Rpl22l1 1.085364e-31 0.5154337 0.984 0.688 4.331146e-27 1 ## Kcnh31 1.106909e-31 1.0015526 0.633 0.224 4.417122e-27 1 ## Rpl18 1.260329e-31 0.4487396 1.000 0.864 5.029343e-27 1 ## Stmn3 1.393263e-31 2.4244497 0.294 0.059 5.559816e-27 1 ## Rprm 1.404663e-31 2.4436707 0.237 0.036 5.605307e-27 1 ## Hint1 1.478598e-31 0.6362327 0.996 0.811 5.900346e-27 1 ## Ndufa1 1.571890e-31 0.6609389 0.992 0.788 6.272626e-27 1 ## Lyplal1 1.628654e-31 1.5895147 0.433 0.124 6.499145e-27 1 ## Fth1 2.000658e-31 0.1854704 1.000 0.953 7.983626e-27 1 ## Rab37 2.403871e-31 1.1031135 0.612 0.215 9.592646e-27 1 ## Ptms 2.778440e-31 0.6064857 0.996 0.871 1.108736e-26 1 ## Hus1 3.140385e-31 1.2297174 0.555 0.197 1.253171e-26 1 ## Il1rap 3.623147e-31 1.3140692 0.445 0.133 1.445817e-26 1 ## Arl10 3.938120e-31 1.0029027 0.657 0.252 1.571507e-26 1 ## Susd4 4.404882e-31 5.3693786 0.098 0.001 1.757768e-26 1 ## Gadd45b1 4.733333e-31 1.1510121 0.788 0.375 1.888837e-26 1 ## C8b 4.948664e-31 2.9789520 0.171 0.016 1.974764e-26 1 ## Pgap1 5.859059e-31 0.9903979 0.776 0.347 2.338057e-26 1 ## Pex19 7.226459e-31 1.0163674 0.767 0.368 2.883718e-26 1 ## Chchd10 8.566930e-31 0.4377800 0.959 0.600 3.418633e-26 1 ## Rgs17 8.812749e-31 0.9868704 0.739 0.334 3.516727e-26 1 ## B4galt4 9.667918e-31 1.3640039 0.494 0.159 3.857982e-26 1 ## Serpina1d1 1.135682e-30 1.0017113 0.384 0.094 4.531939e-26 1 ## Orm3 1.139269e-30 3.9060498 0.114 0.003 4.546251e-26 1 ## Znrf2 1.316385e-30 0.9677801 0.759 0.352 5.253035e-26 1 ## Prdx5 1.532983e-30 0.3877254 0.963 0.715 6.117369e-26 1 ## Mblac2 1.548685e-30 1.7100690 0.412 0.117 6.180027e-26 1 ## Cmas1 1.575247e-30 0.8556826 0.902 0.580 6.286023e-26 1 ## Anp32a 1.604621e-30 0.7750677 0.963 0.568 6.403240e-26 1 ## Sgpl11 1.715765e-30 0.8844165 0.914 0.551 6.846760e-26 1 ## Gm53258 1.895571e-30 1.2558419 0.784 0.428 7.564277e-26 1 ## LOC118568133 2.237205e-30 1.1814737 0.661 0.267 8.927568e-26 1 ## Lst1 2.583749e-30 1.7129580 0.396 0.110 1.031045e-25 1 ## H3f3a1 2.611458e-30 0.6232855 1.000 0.867 1.042102e-25 1 ## 1810037I17Rik 2.611833e-30 0.7329767 0.996 0.751 1.042252e-25 1 ## Pkp1 2.623436e-30 1.7606356 0.416 0.126 1.046882e-25 1 ## C4bp 2.807955e-30 2.6048335 0.155 0.013 1.120514e-25 1 ## Pcca1 2.952307e-30 1.1420240 0.698 0.305 1.178118e-25 1 ## Slc17a4 3.238608e-30 0.8225799 0.873 0.494 1.292367e-25 1 ## Mien11 3.248496e-30 0.7197221 0.971 0.711 1.296312e-25 1 ## Txnip 5.303214e-30 1.1841945 0.878 0.556 2.116248e-25 1 ## Mfsd2a 5.717061e-30 1.0738576 0.620 0.251 2.281393e-25 1 ## Pbld2 5.904745e-30 0.9345909 0.714 0.318 2.356289e-25 1 ## Pebp1 7.729149e-30 0.7290244 0.976 0.714 3.084317e-25 1 ## Isg20 8.382517e-30 1.0229177 0.816 0.423 3.345043e-25 1 ## Cyb5r31 9.839637e-30 0.6183345 0.992 0.812 3.926507e-25 1 ## Rplp0 1.059911e-29 0.1098015 1.000 0.882 4.229575e-25 1 ## Gstm1 1.087379e-29 0.4293148 0.596 0.222 4.339185e-25 1 ## Greb1 1.146668e-29 1.6073627 0.351 0.089 4.575778e-25 1 ## Scg51 1.179929e-29 0.6821394 1.000 0.803 4.708507e-25 1 ## Sdc2 1.367509e-29 1.0863353 0.698 0.310 5.457043e-25 1 ## Gpld1 1.652425e-29 1.5145068 0.433 0.129 6.594002e-25 1 ## Dgkk 1.840394e-29 5.6635229 0.106 0.003 7.344091e-25 1 ## A2ml1 2.025243e-29 0.5878592 0.445 0.129 8.081734e-25 1 ## Lims1 2.504369e-29 0.7696613 0.886 0.472 9.993684e-25 1 ## Ndufb12 2.879216e-29 0.6270651 1.000 0.782 1.148951e-24 1 ## Rpl6 3.106491e-29 0.3241749 0.996 0.893 1.239645e-24 1 ## Ndufb11 3.176441e-29 0.6433786 0.992 0.755 1.267559e-24 1 ## Gprc5c 3.421543e-29 1.0236166 0.624 0.234 1.365367e-24 1 ## Tmem134 3.450541e-29 0.8438280 0.906 0.558 1.376939e-24 1 ## Rasa4 3.747261e-29 0.7821869 0.771 0.341 1.495344e-24 1 ## Hcfc1r1 4.710750e-29 0.8468537 0.902 0.563 1.879825e-24 1 ## Efcab12 4.872145e-29 2.2968257 0.192 0.024 1.944230e-24 1 ## Dap 5.060719e-29 0.9131450 0.861 0.504 2.019480e-24 1 ## Akr1b11 5.734132e-29 0.9924037 0.865 0.479 2.288205e-24 1 ## Plpp3 5.806078e-29 1.6542668 0.351 0.089 2.316915e-24 1 ## Fhit 5.920230e-29 1.3422507 0.469 0.150 2.362468e-24 1 ## Pter 6.356485e-29 1.7338595 0.396 0.115 2.536555e-24 1 ## Plekhg1 7.215973e-29 1.1956393 0.539 0.193 2.879534e-24 1 ## Rpl29 7.775569e-29 0.4276433 0.992 0.788 3.102841e-24 1 ## Rps14 8.743245e-29 0.3620336 1.000 0.894 3.488992e-24 1 ## Rpl7 9.443866e-29 0.2630745 1.000 0.824 3.768575e-24 1 ## Ifnar2 9.570018e-29 1.0818406 0.759 0.393 3.818916e-24 1 ## Man2b21 1.241628e-28 1.1933808 0.641 0.259 4.954716e-24 1 ## Pparg 1.492532e-28 1.9782500 0.265 0.053 5.955951e-24 1 ## Gm34173 1.803896e-28 2.1634535 0.118 0.006 7.198445e-24 1 ## Soat1 1.820406e-28 1.0769289 0.661 0.280 7.264330e-24 1 ## Gm5617 2.310784e-28 1.1292883 0.698 0.331 9.221183e-24 1 ## Galnt7 2.470544e-28 0.8284059 0.857 0.481 9.858707e-24 1 ## Tfpi 2.657641e-28 2.4297490 0.269 0.055 1.060532e-23 1 ## Enpp3 2.684955e-28 0.4597244 0.596 0.224 1.071431e-23 1 ## Tcaf1 2.875898e-28 1.3048143 0.641 0.283 1.147627e-23 1 ## Adgrf5 2.897914e-28 0.4978548 0.784 0.330 1.156412e-23 1 ## Stard4 3.036162e-28 0.8745116 0.731 0.352 1.211580e-23 1 ## Cyb5a 3.301339e-28 0.4734901 0.996 0.714 1.317399e-23 1 ## Rexo2 3.529210e-28 0.9908397 0.755 0.363 1.408331e-23 1 ## Rgs21 3.788106e-28 0.6469651 0.988 0.604 1.511644e-23 1 ## Lypla11 3.905723e-28 0.8606481 0.947 0.736 1.558579e-23 1 ## 1110065P20Rik 4.501247e-28 1.1535944 0.706 0.325 1.796223e-23 1 ## Focad 4.514098e-28 1.5082366 0.424 0.133 1.801351e-23 1 ## Naglu 4.995872e-28 1.2501646 0.478 0.161 1.993603e-23 1 ## Eif1b 5.536652e-28 0.9853312 0.776 0.407 2.209401e-23 1 ## Ctsb 6.635749e-28 0.6760448 0.980 0.734 2.647996e-23 1 ## Slc48a1 6.889355e-28 1.2732199 0.580 0.226 2.749197e-23 1 ## Tmem38b 7.936474e-28 1.4446293 0.433 0.138 3.167050e-23 1 ## Cbr1 8.014905e-28 0.5236981 0.882 0.491 3.198348e-23 1 ## Asl 9.116436e-28 0.8800790 0.718 0.317 3.637914e-23 1 ## Cdkn1c1 1.047948e-27 0.7937464 0.882 0.506 4.181837e-23 1 ## Mrpl331 1.121050e-27 0.7828092 0.955 0.647 4.473551e-23 1 ## Parva 1.194932e-27 1.3718609 0.506 0.181 4.768377e-23 1 ## LOC118567825 1.494270e-27 0.9005741 0.600 0.238 5.962884e-23 1 ## Stx17 1.540184e-27 0.9109458 0.559 0.217 6.146105e-23 1 ## Frrs1 1.631663e-27 0.8749516 0.702 0.313 6.511150e-23 1 ## Tmem97 1.668133e-27 0.9236603 0.767 0.384 6.656684e-23 1 ## Rcan3 2.088184e-27 0.8597072 0.796 0.396 8.332899e-23 1 ## Ctnnb11 2.110644e-27 0.5309419 0.996 0.819 8.422524e-23 1 ## Ffar4 2.246183e-27 0.9870540 0.518 0.178 8.963395e-23 1 ## Coa3 2.447789e-27 0.6528739 0.980 0.672 9.767903e-23 1 ## Tomm20 2.687361e-27 0.6247193 0.959 0.647 1.072391e-22 1 ## Tmem200a1 2.840542e-27 1.2786208 0.392 0.110 1.133518e-22 1 ## 9530068E07Rik1 2.949238e-27 0.8532037 0.914 0.564 1.176894e-22 1 ## Atp6v1f1 3.079105e-27 0.6042606 0.984 0.762 1.228717e-22 1 ## Zfyve21 3.286910e-27 1.0978710 0.563 0.222 1.311642e-22 1 ## Pfdn5 3.349086e-27 0.5449323 0.996 0.813 1.336453e-22 1 ## Tcea3 3.670471e-27 0.9012668 0.473 0.152 1.464701e-22 1 ## Rps8 4.020675e-27 0.1322042 1.000 0.861 1.604450e-22 1 ## Nucb2 5.236952e-27 1.1907413 0.571 0.221 2.089806e-22 1 ## Slc3a11 6.777484e-27 0.7203133 0.898 0.485 2.704555e-22 1 ## Snph 6.799867e-27 1.6324414 0.396 0.121 2.713487e-22 1 ## Eef1b2 7.977069e-27 0.2213611 0.976 0.742 3.183249e-22 1 ## Cox7a21 8.942314e-27 0.4741227 1.000 0.823 3.568430e-22 1 ## Ndrg21 9.760465e-27 1.0745510 0.588 0.229 3.894913e-22 1 ## Sem1 1.254885e-26 0.6170624 0.976 0.718 5.007619e-22 1 ## Sez6l21 1.259129e-26 0.7449230 0.947 0.607 5.024553e-22 1 ## Hps4 1.285790e-26 0.8888015 0.706 0.317 5.130946e-22 1 ## Srp141 1.304395e-26 0.6827853 0.959 0.680 5.205187e-22 1 ## Rpl31 1.341736e-26 0.4620715 0.996 0.760 5.354198e-22 1 ## Hspb6 1.440340e-26 2.6916645 0.163 0.019 5.747678e-22 1 ## Prom11 1.571515e-26 0.9047757 0.657 0.285 6.271129e-22 1 ## Pid1 1.625445e-26 1.8267799 0.306 0.074 6.486337e-22 1 ## Gstt2 1.690068e-26 1.0589988 0.645 0.274 6.744216e-22 1 ## Rapgef3 1.705834e-26 2.5558985 0.159 0.017 6.807131e-22 1 ## Amer2 1.805998e-26 1.9965173 0.249 0.050 7.206833e-22 1 ## Gas5 1.957467e-26 0.4256225 0.955 0.648 7.811271e-22 1 ## Plekha2 2.002012e-26 0.9476048 0.759 0.363 7.989027e-22 1 ## Sat2 2.052756e-26 2.2061876 0.229 0.041 8.191523e-22 1 ## Fas 2.402846e-26 2.2074603 0.220 0.039 9.588555e-22 1 ## Ovol2 2.421475e-26 1.0355503 0.653 0.295 9.662894e-22 1 ## Svil 2.467047e-26 0.7705985 0.682 0.303 9.844753e-22 1 ## Uqcc31 2.559461e-26 0.8886278 0.869 0.477 1.021353e-21 1 ## A730098A19Rik 2.880619e-26 3.6059291 0.127 0.009 1.149511e-21 1 ## Malrd1 3.002043e-26 0.6368010 0.857 0.426 1.197965e-21 1 ## Prps2 3.184006e-26 1.7690450 0.400 0.128 1.270577e-21 1 ## Entpd8 3.307416e-26 0.4972017 0.865 0.464 1.319824e-21 1 ## Tnfaip8l3 3.390410e-26 1.1425144 0.335 0.089 1.352943e-21 1 ## Dut 4.586688e-26 0.7965934 0.665 0.271 1.830318e-21 1 ## Gm20234 4.638282e-26 1.5139576 0.339 0.090 1.850906e-21 1 ## Rps28 4.675600e-26 0.2766217 0.996 0.854 1.865798e-21 1 ## Cfap92 6.714574e-26 2.6674333 0.171 0.022 2.679451e-21 1 ## Pnlip 7.580187e-26 4.6366682 0.082 0.001 3.024874e-21 1 ## Trappc2l 8.664790e-26 0.7464750 0.918 0.556 3.457684e-21 1 ## Rpl30 8.758711e-26 0.3339080 1.000 0.836 3.495164e-21 1 ## Park7 9.811009e-26 0.5998156 0.980 0.679 3.915083e-21 1 ## Slc38a4 1.014844e-25 5.2782355 0.086 0.001 4.049736e-21 1 ## C1d 1.028599e-25 0.7695804 0.853 0.456 4.104623e-21 1 ## Tagln3 1.156534e-25 4.0541776 0.118 0.008 4.615147e-21 1 ## Pmm1 1.437432e-25 1.3945676 0.424 0.143 5.736071e-21 1 ## Nme2 1.786031e-25 0.3547381 0.967 0.697 7.127156e-21 1 ## Ostc 1.901761e-25 0.6502412 0.971 0.690 7.588976e-21 1 ## Apobec3 2.052566e-25 0.8975839 0.657 0.287 8.190765e-21 1 ## Rpl9 2.973383e-25 0.3823235 0.996 0.840 1.186528e-20 1 ## Tspan17 3.110006e-25 1.0230752 0.600 0.259 1.241048e-20 1 ## Imp3 3.266822e-25 0.9009536 0.849 0.488 1.303625e-20 1 ## Mcee 3.488619e-25 0.7755641 0.833 0.419 1.392133e-20 1 ## Psma4 3.657301e-25 0.6055067 0.980 0.690 1.459446e-20 1 ## Rpl8 3.787396e-25 0.2788108 1.000 0.892 1.511360e-20 1 ## Arl6ip5 4.416044e-25 0.9647499 0.718 0.358 1.762222e-20 1 ## Ebp1 4.476541e-25 0.8263282 0.804 0.442 1.786364e-20 1 ## Ccna1 5.274185e-25 2.1626184 0.106 0.006 2.104664e-20 1 ## Pccb 5.576734e-25 0.7232577 0.804 0.410 2.225396e-20 1 ## Arl3 6.047035e-25 0.8869168 0.808 0.432 2.413069e-20 1 ## Arpp191 6.670384e-25 0.5738654 0.992 0.767 2.661817e-20 1 ## Colgalt1 7.327280e-25 1.0071542 0.608 0.256 2.923951e-20 1 ## Rab4b 7.601875e-25 0.7489517 0.849 0.495 3.033528e-20 1 ## Clec2e 7.994899e-25 0.8047492 0.567 0.222 3.190365e-20 1 ## Maob1 8.015717e-25 1.2025923 0.551 0.222 3.198672e-20 1 ## H2az1 8.748503e-25 0.4633283 0.996 0.811 3.491090e-20 1 ## Kdelr1 8.995613e-25 0.7073048 0.939 0.657 3.589699e-20 1 ## Pax6os1 1.023601e-24 1.1758272 0.486 0.169 4.084681e-20 1 ## Ogfod3 1.060765e-24 1.1547622 0.506 0.194 4.232983e-20 1 ## Strip2 1.249003e-24 1.7246826 0.253 0.055 4.984146e-20 1 ## Sult1d1 1.439217e-24 0.5653818 0.947 0.541 5.743197e-20 1 ## Serf2 1.450962e-24 0.3769356 0.996 0.885 5.790064e-20 1 ## Acsl5 1.466330e-24 0.2355850 0.918 0.566 5.851390e-20 1 ## Tenm3 1.581662e-24 1.1793910 0.400 0.125 6.311624e-20 1 ## Rps5 1.702409e-24 0.1414790 1.000 0.835 6.793464e-20 1 ## Tet1 1.796672e-24 1.5092283 0.347 0.099 7.169621e-20 1 ## Vps37d 1.802843e-24 1.6652239 0.343 0.097 7.194244e-20 1 ## Nfib 1.955251e-24 0.8476627 0.543 0.205 7.802427e-20 1 ## Prkacb 2.071357e-24 0.8512254 0.829 0.473 8.265751e-20 1 ## Arhgap31 2.154442e-24 1.0457046 0.429 0.140 8.597301e-20 1 ## Tmem9 2.239675e-24 1.5239843 0.437 0.154 8.937423e-20 1 ## Galnt41 2.290788e-24 0.8045032 0.886 0.556 9.141390e-20 1 ## Hopx1 2.459754e-24 0.5622337 1.000 0.853 9.815647e-20 1 ## Tm4sf1 2.497353e-24 3.7433067 0.090 0.003 9.965686e-20 1 ## Ocrl 2.766038e-24 0.7147813 0.637 0.274 1.103787e-19 1 ## Mmp14 5.147184e-24 0.7153051 0.857 0.512 2.053984e-19 1 ## Cyp4b1 5.586527e-24 0.5718051 0.882 0.520 2.229304e-19 1 ## Ywhaq1 7.281109e-24 0.6448305 0.955 0.672 2.905527e-19 1 ## Dnlz 7.600648e-24 0.8050393 0.649 0.286 3.033038e-19 1 ## Atxn101 7.800847e-24 0.6447303 0.939 0.643 3.112928e-19 1 ## Cdc14a 8.093711e-24 1.2216208 0.433 0.152 3.229796e-19 1 ## Pld3 8.279187e-24 0.9458452 0.682 0.321 3.303810e-19 1 ## Gfra3 8.289541e-24 0.8040290 0.792 0.418 3.307941e-19 1 ## Aplp2 8.534262e-24 0.7947492 0.759 0.377 3.405597e-19 1 ## Tmx11 8.623920e-24 0.7526558 0.890 0.589 3.441375e-19 1 ## Mgat31 8.890721e-24 0.6344787 0.967 0.685 3.547842e-19 1 ## Pgrmc2 9.035210e-24 0.7557658 0.824 0.461 3.605501e-19 1 ## Slc39a2 9.406198e-24 1.7482937 0.265 0.061 3.753543e-19 1 ## Mospd21 1.041226e-23 0.9475249 0.690 0.338 4.155014e-19 1 ## Arhgef38 1.153085e-23 0.5595356 0.980 0.639 4.601385e-19 1 ## Psmb9 1.168559e-23 0.8603215 0.665 0.313 4.663136e-19 1 ## Epha7 1.169546e-23 1.3205947 0.339 0.094 4.667075e-19 1 ## Cela11 1.303095e-23 0.7097356 0.824 0.446 5.200002e-19 1 ## Avil 1.451149e-23 0.5180237 0.212 0.039 5.790812e-19 1 ## Lcmt2 1.727605e-23 1.5422386 0.343 0.102 6.894009e-19 1 ## Fads3 1.783750e-23 1.3672506 0.404 0.134 7.118054e-19 1 ## Ezh2 1.849991e-23 0.8317178 0.669 0.299 7.382390e-19 1 ## Smim6 1.860834e-23 0.6794547 0.959 0.669 7.425658e-19 1 ## Pomgnt2 1.948801e-23 1.6698305 0.302 0.081 7.776692e-19 1 ## Rpl41 2.000190e-23 0.3267598 1.000 0.945 7.981758e-19 1 ## Rab2a1 2.080626e-23 0.5452008 0.992 0.834 8.302738e-19 1 ## Glo1 2.209881e-23 0.7065621 0.845 0.468 8.818529e-19 1 ## Pla2g2f 2.261144e-23 0.7391705 0.890 0.526 9.023094e-19 1 ## Sdf41 2.278622e-23 0.6283545 0.976 0.746 9.092843e-19 1 ## Mexis 2.357660e-23 2.0969511 0.229 0.048 9.408241e-19 1 ## Ntrk2 2.791656e-23 4.0497652 0.094 0.004 1.114010e-18 1 ## Cetn2 2.887439e-23 0.8572092 0.698 0.346 1.152232e-18 1 ## Pdxk1 3.105558e-23 0.6917015 0.906 0.549 1.239273e-18 1 ## Usp18 3.510836e-23 0.8452598 0.653 0.289 1.400999e-18 1 ## Gm29825 3.619128e-23 4.1876889 0.090 0.003 1.444213e-18 1 ## Slc16a11 3.665011e-23 0.6029815 0.776 0.361 1.462523e-18 1 ## Hepacam21 4.046586e-23 0.5439019 1.000 0.875 1.614790e-18 1 ## Zfp4671 4.365737e-23 0.7781056 0.727 0.328 1.742147e-18 1 ## Maged1 4.605638e-23 0.6014004 0.971 0.681 1.837880e-18 1 ## Id21 4.696830e-23 0.6097625 0.976 0.749 1.874270e-18 1 ## Ppib 4.728322e-23 0.5023461 0.984 0.691 1.886837e-18 1 ## Ergic31 4.893850e-23 0.6382721 0.939 0.669 1.952891e-18 1 ## Angptl8 5.021556e-23 1.4859500 0.343 0.100 2.003852e-18 1 ## Col1a2 5.171124e-23 2.5644410 0.086 0.003 2.063537e-18 1 ## Lrpprc 5.177981e-23 1.1135809 0.616 0.286 2.066273e-18 1 ## Tmod1 5.340440e-23 3.5591467 0.106 0.007 2.131102e-18 1 ## Npm1 5.374688e-23 0.2029208 0.988 0.730 2.144769e-18 1 ## Cenpx 6.226014e-23 0.7089386 0.755 0.391 2.484491e-18 1 ## Pecr 7.788756e-23 1.8052703 0.310 0.089 3.108103e-18 1 ## Nxnl2 8.295868e-23 1.3869552 0.314 0.083 3.310466e-18 1 ## Ost4 9.465373e-23 0.6663821 0.886 0.526 3.777157e-18 1 ## Mapk8ip11 9.663563e-23 0.9568054 0.531 0.205 3.856245e-18 1 ## Pitx1 9.751412e-23 0.9572745 0.486 0.192 3.891301e-18 1 ## Slc16a2 1.036580e-22 1.7882378 0.245 0.055 4.136473e-18 1 ## Deptor 1.059130e-22 0.6805212 0.731 0.354 4.226460e-18 1 ## Nit2 1.069869e-22 1.0338859 0.612 0.285 4.269312e-18 1 ## Gm12506 1.107634e-22 2.9009528 0.122 0.011 4.420014e-18 1 ## Cox171 1.148377e-22 0.5103836 0.988 0.785 4.582597e-18 1 ## Morf4l21 1.173426e-22 0.5755428 0.976 0.763 4.682558e-18 1 ## Rps25 1.236041e-22 0.3352146 0.996 0.812 4.932420e-18 1 ## Ap3s11 1.249222e-22 0.7402662 0.873 0.544 4.985022e-18 1 ## Golga7b 1.258595e-22 1.0630534 0.343 0.102 5.022424e-18 1 ## Bmpr1a 1.287270e-22 0.6993146 0.898 0.593 5.136849e-18 1 ## Tmem45b 1.387718e-22 0.7567826 0.829 0.459 5.537690e-18 1 ## Cyba 1.401024e-22 0.4782329 0.694 0.308 5.590786e-18 1 ## Mpc1 1.579389e-22 0.5747417 0.910 0.576 6.302551e-18 1 ## Rack1 1.644790e-22 0.1648713 0.996 0.832 6.563536e-18 1 ## Tmsb101 1.662127e-22 0.4889122 1.000 0.872 6.632716e-18 1 ## Rpl36 1.719125e-22 0.2479952 1.000 0.897 6.860168e-18 1 ## H2-M3 1.723268e-22 1.5431956 0.363 0.117 6.876702e-18 1 ## Slc39a14 1.731709e-22 0.8959578 0.580 0.245 6.910385e-18 1 ## Gm542261 1.997335e-22 2.0093751 0.233 0.050 7.970366e-18 1 ## Pfn2 2.082076e-22 1.2542578 0.404 0.133 8.308524e-18 1 ## Gm34299 2.094725e-22 3.3713868 0.098 0.006 8.359002e-18 1 ## Rpl14 2.113208e-22 0.1968221 0.996 0.829 8.432756e-18 1 ## Calm2 2.180741e-22 0.4864517 0.992 0.841 8.702247e-18 1 ## C1qtnf1 2.214841e-22 2.0342189 0.192 0.034 8.838323e-18 1 ## Cfap36 2.232125e-22 0.8581114 0.743 0.399 8.907293e-18 1 ## Cetn3 2.315896e-22 0.8270070 0.771 0.416 9.241584e-18 1 ## B2m 2.459709e-22 0.2930721 0.996 0.860 9.815468e-18 1 ## Ffar3 2.475495e-22 0.7352232 0.416 0.137 9.878465e-18 1 ## Slc40a1 2.619098e-22 0.5858355 0.380 0.121 1.045151e-17 1 ## Sv2b 2.730198e-22 5.6673059 0.065 0.000 1.089486e-17 1 ## Rheb 2.971472e-22 0.6912046 0.824 0.468 1.185766e-17 1 ## Nudt2 3.091859e-22 1.1737771 0.478 0.186 1.233806e-17 1 ## Prkaa21 3.374982e-22 0.6523804 0.690 0.331 1.346787e-17 1 ## Dync2li1 3.376942e-22 1.2297759 0.461 0.174 1.347569e-17 1 ## Nipsnap3b 3.487830e-22 0.7751590 0.812 0.470 1.391819e-17 1 ## Cbfa2t31 3.691675e-22 0.6704664 0.845 0.463 1.473163e-17 1 ## Rps29 3.734811e-22 0.2269819 1.000 0.904 1.490376e-17 1 ## Bphl 3.783247e-22 1.0673414 0.608 0.285 1.509705e-17 1 ## Fkbp1a1 4.078560e-22 0.5180536 0.996 0.797 1.627549e-17 1 ## Cpd1 4.248095e-22 0.7078203 0.922 0.594 1.695203e-17 1 ## Ppp1cb1 4.251839e-22 0.4839204 0.963 0.771 1.696696e-17 1 ## Stxbp6 4.917512e-22 1.5695163 0.302 0.085 1.962333e-17 1 ## Tax1bp1 4.940712e-22 0.4812055 0.996 0.890 1.971591e-17 1 ## Man2b1 5.274387e-22 0.8102647 0.722 0.385 2.104744e-17 1 ## Nfic 5.711545e-22 0.8441935 0.645 0.289 2.279192e-17 1 ## Foxa2 6.645530e-22 0.8745997 0.714 0.365 2.651899e-17 1 ## Stard3nl 6.999162e-22 1.0331860 0.608 0.291 2.793016e-17 1 ## Gm52543 7.435423e-22 2.8859878 0.139 0.017 2.967106e-17 1 ## Itga21 7.573820e-22 0.7839585 0.527 0.211 3.022333e-17 1 ## Tubb2b1 8.990524e-22 0.5443773 0.849 0.444 3.587669e-17 1 ## Chst8 9.344306e-22 2.0616327 0.286 0.078 3.728845e-17 1 ## Ntan1 9.967196e-22 0.7677777 0.816 0.440 3.977410e-17 1 ## Ccdc50 1.014514e-21 0.6628339 0.796 0.470 4.048417e-17 1 ## Zfp30 1.213390e-21 1.1866708 0.318 0.091 4.842032e-17 1 ## Zbed3 1.343837e-21 1.4289167 0.347 0.112 5.362581e-17 1 ## Fat4 1.357067e-21 1.5678476 0.212 0.043 5.415377e-17 1 ## Nucb1 1.398981e-21 0.6232764 0.976 0.692 5.582633e-17 1 ## Pde1c 1.433704e-21 1.0603056 0.433 0.155 5.721196e-17 1 ## Psen21 1.599106e-21 0.8880808 0.710 0.359 6.381232e-17 1 ## Tmem38a 1.704246e-21 0.8388644 0.510 0.200 6.800794e-17 1 ## Rpl21 1.710821e-21 0.3109734 1.000 0.865 6.827032e-17 1 ## Gp1bb 1.793686e-21 1.1922840 0.478 0.199 7.157703e-17 1 ## Hmgb1 1.880861e-21 0.2869453 0.988 0.809 7.505574e-17 1 ## Ggh 1.955205e-21 0.8397844 0.657 0.314 7.802244e-17 1 ## Septin5 1.991417e-21 1.2769337 0.420 0.157 7.946750e-17 1 ## Prdx21 2.071336e-21 0.5358139 0.992 0.778 8.265666e-17 1 ## Plaat3 2.135854e-21 0.6842951 0.527 0.215 8.523124e-17 1 ## Rab39b 2.199739e-21 1.2282932 0.298 0.081 8.778059e-17 1 ## Aplp11 2.508064e-21 0.4976444 0.992 0.807 1.000843e-16 1 ## Raph1 2.533664e-21 0.4847912 0.922 0.619 1.011059e-16 1 ## Hmgn5 2.545772e-21 0.6580523 0.776 0.383 1.015890e-16 1 ## Tmem2561 2.649857e-21 0.5716754 0.955 0.649 1.057425e-16 1 ## Ceacam18 2.758648e-21 0.7684128 0.731 0.365 1.100838e-16 1 ## Gm5148 2.826296e-21 3.4497267 0.106 0.008 1.127833e-16 1 ## Steap2 2.854403e-21 0.8643967 0.510 0.212 1.139049e-16 1 ## Chd6 2.857815e-21 0.5683043 0.865 0.509 1.140411e-16 1 ## Tspan131 2.922775e-21 0.5446731 0.992 0.775 1.166333e-16 1 ## Rpl7a 2.935692e-21 0.2492600 1.000 0.830 1.171488e-16 1 ## Hsd17b13 3.097265e-21 2.0946230 0.261 0.070 1.235964e-16 1 ## 0610040J01Rik 3.167319e-21 0.8629502 0.702 0.367 1.263919e-16 1 ## Slc35b3 3.305125e-21 1.0175161 0.543 0.240 1.318910e-16 1 ## Pde8a1 3.753764e-21 0.7600621 0.600 0.261 1.497940e-16 1 ## Dnajc15 4.386825e-21 0.7031568 0.657 0.296 1.750562e-16 1 ## Cox8a1 4.617831e-21 0.4401843 0.996 0.866 1.842746e-16 1 ## Fah 4.736406e-21 1.8864020 0.200 0.040 1.890063e-16 1 ## Arpc1a1 4.877665e-21 0.5657519 0.951 0.687 1.946432e-16 1 ## Mrpl17 5.088173e-21 0.9704557 0.751 0.391 2.030435e-16 1 ## Gm32995 5.511484e-21 5.3407265 0.061 0.000 2.199358e-16 1 ## Samd11 6.045426e-21 1.2151481 0.204 0.041 2.412427e-16 1 ## Ap1s3 6.276857e-21 1.0177818 0.371 0.128 2.504780e-16 1 ## Immp1l 6.726295e-21 0.7999565 0.714 0.371 2.684128e-16 1 ## Pck2 7.165452e-21 0.9272985 0.543 0.222 2.859374e-16 1 ## Galr2 7.218659e-21 2.4554474 0.131 0.015 2.880606e-16 1 ## Gm30247 7.501148e-21 1.3940729 0.200 0.040 2.993333e-16 1 ## Ern1 7.535437e-21 0.6549997 0.665 0.300 3.007016e-16 1 ## Kank2 8.218102e-21 1.3430928 0.286 0.078 3.279433e-16 1 ## Epha31 8.270664e-21 2.2745326 0.163 0.026 3.300408e-16 1 ## Sec61g1 9.235235e-21 0.4754915 1.000 0.814 3.685321e-16 1 ## Ap1ar 1.016710e-20 0.5850322 0.853 0.528 4.057183e-16 1 ## Pgm2l1 1.027879e-20 1.4208007 0.371 0.132 4.101752e-16 1 ## Eef1g 1.057741e-20 0.3235808 0.988 0.744 4.220916e-16 1 ## Iftap 1.143845e-20 1.0862895 0.396 0.143 4.564512e-16 1 ## Cobll1 1.156657e-20 1.0229839 0.543 0.240 4.615640e-16 1 ## Fam204a 1.351130e-20 0.6322951 0.776 0.408 5.391685e-16 1 ## Sec11a1 1.375874e-20 0.6398308 0.939 0.655 5.490425e-16 1 ## Psma5 1.482783e-20 0.5845686 0.853 0.528 5.917045e-16 1 ## Nagk 1.506129e-20 0.7179928 0.878 0.544 6.010208e-16 1 ## Erg28 1.540383e-20 0.8727809 0.743 0.400 6.146899e-16 1 ## Tmem45a 1.615058e-20 4.7178269 0.073 0.002 6.444890e-16 1 ## Mthfs 1.615507e-20 1.5162061 0.380 0.136 6.446680e-16 1 ## Arhgef37 1.670494e-20 1.4538982 0.233 0.054 6.666108e-16 1 ## Mir100hg 1.780334e-20 4.7784747 0.073 0.002 7.104423e-16 1 ## Nbea 1.853192e-20 0.5346812 0.959 0.724 7.395161e-16 1 ## Gask1a 1.975995e-20 3.8740628 0.073 0.002 7.885206e-16 1 ## Manf 2.004894e-20 0.6123468 0.963 0.674 8.000530e-16 1 ## Vmac 2.033737e-20 1.4974678 0.367 0.126 8.115629e-16 1 ## Tecr1 2.146213e-20 0.5471068 0.992 0.769 8.564462e-16 1 ## Caprin2 2.227512e-20 2.1578085 0.176 0.032 8.888886e-16 1 ## Pank1 2.350268e-20 1.1105859 0.457 0.184 9.378744e-16 1 ## Ormdl21 2.401587e-20 0.8010923 0.780 0.444 9.583535e-16 1 ## Rab27a 2.410149e-20 1.3045123 0.371 0.129 9.617701e-16 1 ## Pla2g15 2.582090e-20 1.9509467 0.224 0.052 1.030383e-15 1 ## Serf1 2.672648e-20 0.8976876 0.698 0.349 1.066520e-15 1 ## Pigp 2.831992e-20 0.8196960 0.829 0.491 1.130106e-15 1 ## Sfr1 2.898499e-20 0.5207521 0.947 0.644 1.156646e-15 1 ## Frmd7 2.929716e-20 3.0543001 0.094 0.006 1.169103e-15 1 ## Selenoi 3.105187e-20 0.7033199 0.600 0.284 1.239125e-15 1 ## Rida 3.328903e-20 1.0110673 0.539 0.235 1.328399e-15 1 ## Creg11 3.336952e-20 0.6651022 0.894 0.548 1.331611e-15 1 ## Adam32 3.662564e-20 3.3747673 0.090 0.006 1.461546e-15 1 ## Rps9 3.692682e-20 0.1890479 1.000 0.875 1.473565e-15 1 ## Rab11fip2 3.800292e-20 0.7762609 0.641 0.305 1.516507e-15 1 ## Gsto2 3.968447e-20 1.3548760 0.306 0.092 1.583609e-15 1 ## Nav2 4.222893e-20 0.9524607 0.665 0.333 1.685146e-15 1 ## Rpl24 4.375055e-20 0.2205654 1.000 0.887 1.745866e-15 1 ## Mxd4 4.756580e-20 0.5333989 0.935 0.621 1.898113e-15 1 ## Gm2a 4.820428e-20 1.5325146 0.318 0.105 1.923592e-15 1 ## Per3 5.512312e-20 1.0199112 0.392 0.145 2.199688e-15 1 ## Calm31 5.629532e-20 0.4496101 1.000 0.818 2.246465e-15 1 ## Lsr1 6.261443e-20 0.5578103 0.984 0.783 2.498629e-15 1 ## Hpgd 6.338314e-20 0.7049549 0.751 0.419 2.529304e-15 1 ## Lamb1 6.629928e-20 2.7594166 0.127 0.015 2.645673e-15 1 ## Selenow 6.914407e-20 0.6455501 0.939 0.712 2.759194e-15 1 ## Ppdpf1 7.327734e-20 0.5618664 0.939 0.654 2.924132e-15 1 ## Trappc6b1 7.800723e-20 0.5962409 0.914 0.583 3.112878e-15 1 ## Sobp1 8.130364e-20 0.6931675 0.633 0.290 3.244422e-15 1 ## Lamtor21 9.012763e-20 0.5520689 0.947 0.711 3.596543e-15 1 ## Sp3os1 9.585351e-20 0.7926916 0.653 0.328 3.825034e-15 1 ## 2510009E07Rik 1.052510e-19 0.9460376 0.535 0.233 4.200043e-15 1 ## Ifit1 1.069444e-19 0.5965519 0.347 0.113 4.267615e-15 1 ## Antkmt1 1.107128e-19 0.5702952 0.935 0.613 4.417996e-15 1 ## Slc35g21 1.124300e-19 0.8085909 0.731 0.386 4.486518e-15 1 ## Rpl18a 1.191599e-19 0.1016208 1.000 0.857 4.755078e-15 1 ## Agtpbp1 1.204159e-19 0.9749456 0.404 0.147 4.805198e-15 1 ## Glrx5 1.219118e-19 0.5044110 0.837 0.449 4.864891e-15 1 ## Prkrip11 1.250461e-19 0.7447285 0.686 0.349 4.989967e-15 1 ## Guk11 1.286793e-19 0.5217005 0.984 0.725 5.134948e-15 1 ## Med12 1.318632e-19 0.8270937 0.527 0.230 5.262000e-15 1 ## Stxbp11 1.374127e-19 0.6183843 0.865 0.512 5.483453e-15 1 ## Ndufa71 1.444286e-19 0.5550983 0.984 0.750 5.763422e-15 1 ## Btf31 1.570275e-19 0.3481238 0.988 0.789 6.266183e-15 1 ## Ccdc169 1.630499e-19 3.4434685 0.082 0.004 6.506506e-15 1 ## Ehd3 1.964328e-19 2.6015784 0.110 0.011 7.838649e-15 1 ## Cd99l2 2.056722e-19 1.1461411 0.371 0.131 8.207347e-15 1 ## Mturn 2.149642e-19 1.1221188 0.339 0.110 8.578146e-15 1 ## Abcc5 2.169905e-19 0.5626418 0.735 0.399 8.659007e-15 1 ## Krtcap2 2.281521e-19 0.4888626 0.963 0.676 9.104408e-15 1 ## P3h4 2.323236e-19 1.5828461 0.310 0.096 9.270872e-15 1 ## Smim27 2.541294e-19 0.9629034 0.527 0.233 1.014104e-14 1 ## Tbca 2.574411e-19 0.5775682 0.959 0.662 1.027319e-14 1 ## Chst12 2.648408e-19 3.9897662 0.090 0.006 1.056847e-14 1 ## Cul1 2.686054e-19 0.5522064 0.927 0.572 1.071870e-14 1 ## Rplp2 3.252593e-19 0.1260696 1.000 0.854 1.297947e-14 1 ## Spp2 3.424706e-19 2.6988033 0.114 0.013 1.366629e-14 1 ## Gm34194 3.551212e-19 2.3229218 0.131 0.018 1.417111e-14 1 ## Cdhr2 3.625604e-19 0.3586810 0.988 0.841 1.446797e-14 1 ## Krtcap3 3.955223e-19 0.6297095 0.882 0.539 1.578332e-14 1 ## Ten1 3.994098e-19 0.6182382 0.865 0.549 1.593845e-14 1 ## Isoc2b 4.003277e-19 1.0015041 0.490 0.212 1.597508e-14 1 ## Ms4a5 4.038319e-19 2.4147996 0.090 0.006 1.611491e-14 1 ## Eci2 4.679187e-19 0.6403350 0.853 0.503 1.867230e-14 1 ## Rps21 4.730995e-19 0.2193677 1.000 0.858 1.887904e-14 1 ## Gm41676 4.943823e-19 2.4754950 0.122 0.015 1.972833e-14 1 ## Asxl3 5.078976e-19 1.8629612 0.196 0.043 2.026765e-14 1 ## 2610035D17Rik1 5.609329e-19 0.8076294 0.673 0.345 2.238403e-14 1 ## Crybb1 5.727781e-19 0.5807453 0.412 0.144 2.285671e-14 1 ## Fndc1 5.730728e-19 2.2801671 0.098 0.008 2.286847e-14 1 ## Slc44a3 6.571801e-19 0.6697500 0.800 0.451 2.622477e-14 1 ## Peg131 7.072273e-19 0.8104775 0.608 0.287 2.822190e-14 1 ## Cnot8 7.213471e-19 0.7431246 0.571 0.273 2.878536e-14 1 ## Actn11 7.310189e-19 0.4741697 0.971 0.757 2.917131e-14 1 ## Zfp697 7.360352e-19 1.0619654 0.314 0.099 2.937148e-14 1 ## Tmx4 7.913062e-19 0.9031619 0.592 0.266 3.157707e-14 1 ## Nfia 7.993670e-19 0.7884867 0.559 0.258 3.189874e-14 1 ## Tmem107 8.265462e-19 1.1949679 0.367 0.133 3.298333e-14 1 ## Tmem30b1 8.442527e-19 0.6668030 0.849 0.537 3.368991e-14 1 ## Mapk8ip2 8.808228e-19 0.7571103 0.514 0.215 3.514923e-14 1 ## Jade2 8.930331e-19 1.8140537 0.241 0.066 3.563649e-14 1 ## Ceacam2 9.432241e-19 3.3870632 0.102 0.010 3.763936e-14 1 ## Flot1 9.471503e-19 0.7715809 0.616 0.313 3.779603e-14 1 ## Ndufa5 9.542193e-19 0.3988602 0.967 0.688 3.807812e-14 1 ## Gipc1 9.969488e-19 0.8597023 0.588 0.273 3.978324e-14 1 ## Ssr2 1.029805e-18 0.5448539 0.984 0.718 4.109438e-14 1 ## Tpm41 1.067086e-18 0.6447606 0.857 0.593 4.258208e-14 1 ## Tdrkh 1.097190e-18 1.7520024 0.229 0.057 4.378337e-14 1 ## Dynlt31 1.107349e-18 0.5787865 0.898 0.542 4.418878e-14 1 ## Npdc11 1.177041e-18 0.5463312 0.988 0.734 4.696983e-14 1 ## Uqcr111 1.214635e-18 0.3635324 1.000 0.823 4.847001e-14 1 ## Ncbp2as2 1.253387e-18 0.5307027 0.849 0.518 5.001640e-14 1 ## Tm4sf51 1.275017e-18 0.3174110 0.996 0.843 5.087954e-14 1 ## Fech 1.298253e-18 0.6638950 0.633 0.304 5.180680e-14 1 ## Mif1 1.343297e-18 0.2796584 0.902 0.534 5.360426e-14 1 ## Unc80 1.444164e-18 0.4836364 0.518 0.216 5.762936e-14 1 ## Nipal1 1.457346e-18 0.6509157 0.563 0.260 5.815539e-14 1 ## Cxxc51 1.590643e-18 0.6095212 0.869 0.501 6.347461e-14 1 ## Galns 1.594651e-18 1.4084374 0.339 0.122 6.363454e-14 1 ## Pon3 1.617162e-18 0.5937262 0.878 0.561 6.453286e-14 1 ## Appl1 1.636300e-18 0.5485234 0.735 0.385 6.529655e-14 1 ## Klf2 2.015779e-18 1.2494287 0.437 0.179 8.043965e-14 1 ## Unc5c 2.237243e-18 5.7196419 0.053 0.000 8.927719e-14 1 ## Cops91 2.262380e-18 0.5180576 0.947 0.690 9.028029e-14 1 ## Prdx6 2.385882e-18 0.2873314 0.788 0.429 9.520861e-14 1 ## Map1b 2.505775e-18 0.4257414 0.980 0.725 9.999296e-14 1 ## Tbrg1 2.603757e-18 0.5146462 0.988 0.690 1.039029e-13 1 ## Kif5b1 2.608598e-18 0.4126073 0.996 0.899 1.040961e-13 1 ## Hexa1 2.627435e-18 0.5620841 0.918 0.606 1.048478e-13 1 ## Mamld1 2.748763e-18 2.2017615 0.196 0.045 1.096894e-13 1 ## Dnajc121 2.778275e-18 0.6207863 0.869 0.516 1.108671e-13 1 ## Saraf 2.828632e-18 0.4666096 0.971 0.704 1.128766e-13 1 ## Ppp1r14b 3.008927e-18 0.5074638 0.780 0.424 1.200712e-13 1 ## Cbx6 3.028411e-18 0.5742870 0.780 0.433 1.208487e-13 1 ## Rabac11 3.030323e-18 0.6594585 0.865 0.569 1.209250e-13 1 ## Ndufs51 3.042790e-18 0.5029698 0.980 0.735 1.214225e-13 1 ## Fgf1 3.119427e-18 2.3434937 0.151 0.027 1.244807e-13 1 ## Gm31471 3.287392e-18 1.4324824 0.224 0.056 1.311834e-13 1 ## Jpt2 3.289100e-18 1.1671946 0.408 0.164 1.312515e-13 1 ## Esd 3.615351e-18 0.3879973 0.951 0.656 1.442706e-13 1 ## Ppa1 3.735664e-18 0.3043784 0.951 0.630 1.490717e-13 1 ## Map2k6 3.744732e-18 0.9087278 0.539 0.259 1.494335e-13 1 ## Casp1 4.020217e-18 0.5741293 0.596 0.271 1.604268e-13 1 ## Cebpzos 4.092041e-18 0.7358794 0.722 0.384 1.632929e-13 1 ## Dpm31 4.128803e-18 0.5820708 0.910 0.598 1.647599e-13 1 ## Txndc15 4.184717e-18 0.8888274 0.665 0.354 1.669911e-13 1 ## Elavl4 4.249666e-18 2.7428315 0.135 0.021 1.695829e-13 1 ## Xkr7 4.316937e-18 1.1655820 0.351 0.120 1.722674e-13 1 ## Hes61 4.575757e-18 0.5360005 0.873 0.566 1.825956e-13 1 ## Rnasek1 4.645436e-18 0.4741802 0.976 0.767 1.853761e-13 1 ## Rps26 4.960634e-18 0.2518087 1.000 0.864 1.979541e-13 1 ## Pdk31 5.092425e-18 0.5860618 0.902 0.572 2.032132e-13 1 ## C230057M02Rik 5.098509e-18 3.9599742 0.069 0.003 2.034560e-13 1 ## Ybx1 5.190792e-18 0.2088027 0.992 0.837 2.071385e-13 1 ## Lamc2 5.472843e-18 1.3245917 0.245 0.067 2.183938e-13 1 ## Ntn4 5.867098e-18 1.4359677 0.208 0.050 2.341266e-13 1 ## C330008A17Rik 5.959688e-18 3.2426915 0.094 0.008 2.378213e-13 1 ## Asap3 5.994678e-18 1.5147497 0.233 0.061 2.392176e-13 1 ## Garin5a 6.084035e-18 1.5519312 0.224 0.057 2.427834e-13 1 ## Hsbp11 6.195053e-18 0.4890565 0.947 0.710 2.472136e-13 1 ## Kif12 6.376027e-18 0.6553802 0.743 0.397 2.544354e-13 1 ## Gm132031 6.481998e-18 1.1648892 0.384 0.144 2.586641e-13 1 ## Gc 6.503610e-18 3.4837002 0.061 0.001 2.595266e-13 1 ## Psip1 6.562899e-18 0.7218525 0.816 0.472 2.618925e-13 1 ## Npc2 6.877940e-18 0.5724615 0.943 0.651 2.744642e-13 1 ## Cpt2 7.064780e-18 0.5349658 0.849 0.509 2.819200e-13 1 ## Chchd4 7.116951e-18 0.6984095 0.584 0.266 2.840019e-13 1 ## Pgls 7.155855e-18 0.5478089 0.939 0.630 2.855544e-13 1 ## Xist 7.870441e-18 0.2211388 1.000 0.929 3.140699e-13 1 ## Serpinb9 8.365385e-18 1.3473374 0.351 0.128 3.338207e-13 1 ## Alg5 8.723949e-18 0.6832611 0.718 0.390 3.481292e-13 1 ## Pgghg 8.757985e-18 1.0505634 0.306 0.101 3.494874e-13 1 ## Xkr6 8.832315e-18 0.5003419 0.551 0.245 3.524535e-13 1 ## Atraid 8.990218e-18 0.6708026 0.796 0.475 3.587547e-13 1 ## Uqcc21 9.054536e-18 0.4708857 0.980 0.713 3.613213e-13 1 ## Ttc39b1 9.087766e-18 0.6722328 0.690 0.361 3.626473e-13 1 ## Tmem191 9.529675e-18 1.2461235 0.302 0.096 3.802817e-13 1 ## Higd2a1 9.954266e-18 0.5801830 0.910 0.573 3.972250e-13 1 ## Amigo1 1.019133e-17 0.9550370 0.478 0.203 4.066852e-13 1 ## Gfpt1 1.028035e-17 0.5386951 0.902 0.646 4.102374e-13 1 ## Lingo1 1.219709e-17 1.3109740 0.245 0.068 4.867248e-13 1 ## 3110040N11Rik 1.229490e-17 0.8074718 0.555 0.247 4.906280e-13 1 ## Eif1ax 1.243155e-17 0.6248971 0.755 0.411 4.960811e-13 1 ## Kcnh2 1.256112e-17 1.0922349 0.331 0.112 5.012515e-13 1 ## Hdac5 1.280316e-17 0.6881370 0.624 0.301 5.109100e-13 1 ## Mrfap11 1.284246e-17 0.4689672 0.984 0.794 5.124785e-13 1 ## Frk 1.286329e-17 0.5157664 0.796 0.456 5.133095e-13 1 ## Gm53610 1.385004e-17 1.2293404 0.253 0.072 5.526860e-13 1 ## Mrpl41 1.423956e-17 0.6860625 0.686 0.346 5.682294e-13 1 ## Ggct 1.453688e-17 1.4789826 0.290 0.098 5.800944e-13 1 ## Tusc31 1.506005e-17 0.5524214 0.955 0.698 6.009714e-13 1 ## 2900076A07Rik 1.527926e-17 0.7630494 0.429 0.172 6.097190e-13 1 ## Dhx32 1.567530e-17 0.5033727 0.800 0.453 6.255230e-13 1 ## Fam114a1 1.672315e-17 0.7140115 0.612 0.295 6.673371e-13 1 ## Vdac3 1.790344e-17 0.4855464 0.906 0.585 7.144367e-13 1 ## Tiparp 1.810527e-17 0.8045835 0.633 0.332 7.224909e-13 1 ## Dph6 1.870901e-17 1.0307259 0.416 0.175 7.465832e-13 1 ## Idnk 1.893180e-17 0.6092594 0.820 0.488 7.554736e-13 1 ## Tmem71 1.925089e-17 1.3852009 0.188 0.041 7.682069e-13 1 ## Ypel2 2.150805e-17 0.5217208 0.914 0.645 8.582786e-13 1 ## Sec61b 2.259393e-17 0.4866525 0.992 0.737 9.016108e-13 1 ## Gm33502 2.416284e-17 2.5867264 0.114 0.015 9.642181e-13 1 ## Commd7 2.528248e-17 0.7165840 0.767 0.454 1.008897e-12 1 ## Ppm1l 2.653491e-17 1.0312579 0.429 0.179 1.058876e-12 1 ## Ooep 2.885628e-17 3.4455889 0.086 0.007 1.151510e-12 1 ## Smim14 3.176334e-17 0.5213060 0.906 0.610 1.267516e-12 1 ## Ddb11 3.255384e-17 0.4550522 0.967 0.692 1.299061e-12 1 ## Hmgn2 3.528623e-17 0.5156279 0.878 0.533 1.408097e-12 1 ## Ifi47 3.533864e-17 1.2551314 0.355 0.135 1.410188e-12 1 ## Clic51 3.572705e-17 0.4381720 0.776 0.440 1.425688e-12 1 ## Hap1 3.588387e-17 0.5681896 0.657 0.319 1.431946e-12 1 ## Akr1e1 3.673115e-17 0.8243891 0.551 0.266 1.465757e-12 1 ## Nedd81 3.745863e-17 0.4473297 0.971 0.718 1.494787e-12 1 ## Spmip3 3.837668e-17 1.4009628 0.241 0.069 1.531421e-12 1 ## Limd2 3.852446e-17 0.9672185 0.302 0.099 1.537319e-12 1 ## Rps17 4.037000e-17 0.1842308 0.996 0.808 1.610965e-12 1 ## Gfra2 4.501813e-17 5.1854767 0.049 0.000 1.796448e-12 1 ## Golm2 4.512993e-17 1.0280204 0.404 0.160 1.800910e-12 1 ## Kctd10 4.548079e-17 0.9903566 0.486 0.218 1.814911e-12 1 ## Celf6 4.789324e-17 0.8065156 0.367 0.136 1.911180e-12 1 ## Inpp1 4.822936e-17 0.7555923 0.531 0.244 1.924593e-12 1 ## Sytl5 5.000580e-17 0.4418782 0.698 0.350 1.995481e-12 1 ## Dnajc18 5.004437e-17 0.9250695 0.469 0.202 1.997021e-12 1 ## Tmem266 5.119355e-17 2.6740070 0.151 0.029 2.042879e-12 1 ## Gm40109 5.206016e-17 4.1791795 0.069 0.003 2.077461e-12 1 ## Cep85 5.252892e-17 0.9884091 0.351 0.129 2.096167e-12 1 ## Dram2 5.364948e-17 0.6149071 0.804 0.472 2.140883e-12 1 ## Sec621 5.379329e-17 0.4538016 0.984 0.780 2.146621e-12 1 ## Kras1 5.540927e-17 0.4802963 0.898 0.641 2.211107e-12 1 ## Tmtc4 5.909237e-17 1.0685661 0.367 0.145 2.358081e-12 1 ## Spcs1 5.918706e-17 0.5283787 0.931 0.637 2.361860e-12 1 ## Ndufc2 6.210670e-17 0.4097407 0.959 0.672 2.478368e-12 1 ## Ntaq1 6.801837e-17 1.1359044 0.396 0.157 2.714273e-12 1 ## Mtfp1 6.932335e-17 0.8825441 0.551 0.268 2.766348e-12 1 ## Mapk15 6.949289e-17 1.0387712 0.290 0.093 2.773114e-12 1 ## Evl 7.959350e-17 0.8325932 0.473 0.201 3.176179e-12 1 ## Syp1 8.088434e-17 0.5442568 0.894 0.579 3.227690e-12 1 ## Fdps1 8.175898e-17 0.5371939 0.898 0.604 3.262592e-12 1 ## Flywch21 8.439883e-17 0.8090931 0.596 0.288 3.367935e-12 1 ## Dap3 8.541038e-17 0.5476650 0.816 0.453 3.408301e-12 1 ## Ndufa131 8.681676e-17 0.3986211 0.992 0.839 3.464423e-12 1 ## Rps27l1 8.772393e-17 0.3369916 0.988 0.773 3.500623e-12 1 ## Acbd6 9.058087e-17 0.7843175 0.690 0.370 3.614630e-12 1 ## Gcnt2 9.684116e-17 0.6548289 0.649 0.328 3.864446e-12 1 ## Mast2 1.018488e-16 0.4395955 0.927 0.634 4.064274e-12 1 ## Gm35835 1.139610e-16 4.1693566 0.061 0.002 4.547613e-12 1 ## Gja10 1.155044e-16 3.8783108 0.061 0.002 4.609203e-12 1 ## Ndufb21 1.174998e-16 0.4491897 0.988 0.740 4.688831e-12 1 ## Borcs71 1.184090e-16 0.7481228 0.714 0.400 4.725109e-12 1 ## Snhg8 1.202376e-16 0.6654408 0.669 0.349 4.798081e-12 1 ## Celf4 1.212410e-16 1.3569444 0.200 0.049 4.838124e-12 1 ## Zfp6671 1.214695e-16 0.6447883 0.596 0.287 4.847239e-12 1 ## Gm41964 1.249405e-16 4.0283136 0.057 0.001 4.985752e-12 1 ## Dusp6 1.267156e-16 0.6360650 0.551 0.265 5.056588e-12 1 ## Klhdc8a1 1.363101e-16 1.2868743 0.241 0.068 5.439456e-12 1 ## Neurl1a 1.439669e-16 1.0870973 0.282 0.089 5.745000e-12 1 ## Ube2s 1.477665e-16 0.4439790 0.922 0.632 5.896622e-12 1 ## Eif3k1 1.487189e-16 0.4603962 0.947 0.674 5.934626e-12 1 ## Selenof 1.494372e-16 0.5032067 0.963 0.706 5.963290e-12 1 ## Sstr51 1.509428e-16 0.5281432 0.678 0.328 6.023371e-12 1 ## Zfp507 1.521616e-16 0.9452465 0.404 0.161 6.072007e-12 1 ## Tiprl 1.583737e-16 0.6032119 0.710 0.377 6.319902e-12 1 ## Fam234b 1.596475e-16 0.7074553 0.539 0.254 6.370732e-12 1 ## Pigs1 1.623007e-16 0.7581792 0.624 0.324 6.476610e-12 1 ## 2010004M13Rik 1.674936e-16 1.0372023 0.424 0.176 6.683830e-12 1 ## Mzt2 1.684688e-16 0.9640040 0.457 0.204 6.722749e-12 1 ## Copa1 1.868976e-16 0.4205284 0.959 0.726 7.458150e-12 1 ## Zfp9381 1.923340e-16 0.7865187 0.563 0.271 7.675088e-12 1 ## Lage3 2.004723e-16 0.8911188 0.637 0.337 7.999849e-12 1 ## Rb1 2.217579e-16 0.4422342 0.747 0.412 8.849250e-12 1 ## Gm39467 2.313778e-16 0.4958264 0.845 0.528 9.233133e-12 1 ## Gm9908 2.347847e-16 0.7843643 0.412 0.166 9.369082e-12 1 ## Gstm6 2.350603e-16 0.9323396 0.473 0.203 9.380083e-12 1 ## Pla2g1b 2.355047e-16 1.0807433 0.143 0.027 9.397814e-12 1 ## Rpl5 2.377777e-16 0.1980646 0.996 0.830 9.488519e-12 1 ## Camk1 2.433969e-16 1.3507423 0.318 0.117 9.712753e-12 1 ## Ina 2.439963e-16 0.9408475 0.233 0.067 9.736671e-12 1 ## Pitpnc1 2.473468e-16 0.8203071 0.331 0.115 9.870376e-12 1 ## Dctn31 2.709500e-16 0.4579999 0.984 0.715 1.081226e-11 1 ## Arpc31 2.823883e-16 0.4205735 0.988 0.739 1.126871e-11 1 ## Gmpr 2.846900e-16 0.9024332 0.449 0.194 1.136055e-11 1 ## Psenen1 2.865106e-16 0.4567136 0.976 0.751 1.143321e-11 1 ## Lppos 3.418737e-16 0.6516174 0.612 0.328 1.364247e-11 1 ## Ugt2b35 3.552726e-16 1.2830507 0.224 0.066 1.417715e-11 1 ## Tmem591 3.636138e-16 0.4284133 0.992 0.839 1.451001e-11 1 ## Atp5f1b 3.654165e-16 0.1682823 1.000 0.891 1.458194e-11 1 ## Kxd1 3.716086e-16 0.6098522 0.608 0.309 1.482904e-11 1 ## Akr1c19 3.789002e-16 0.6954340 0.637 0.328 1.512001e-11 1 ## Adgrg41 3.804402e-16 0.4603181 0.988 0.779 1.518147e-11 1 ## Gm41804 3.839141e-16 0.4173332 0.527 0.242 1.532009e-11 1 ## Gm9926 3.873457e-16 0.4739603 0.776 0.449 1.545703e-11 1 ## Mthfsl1 3.971233e-16 0.6993540 0.678 0.396 1.584721e-11 1 ## Fam241b 4.025913e-16 0.7331391 0.535 0.252 1.606541e-11 1 ## Tns2 4.056302e-16 1.0342539 0.278 0.089 1.618667e-11 1 ## Ifi301 4.430908e-16 0.4400704 0.967 0.706 1.768154e-11 1 ## Ruvbl1 4.456499e-16 1.0201927 0.449 0.198 1.778366e-11 1 ## Wdr83os 4.463102e-16 0.6855675 0.808 0.473 1.781001e-11 1 ## Zfp945 4.563970e-16 0.7813222 0.510 0.239 1.821252e-11 1 ## Gng101 4.626142e-16 0.5252666 0.710 0.379 1.846062e-11 1 ## Tnfsf12 4.735959e-16 1.7362016 0.188 0.045 1.889885e-11 1 ## Wfdc2 4.800760e-16 0.7607372 0.380 0.157 1.915743e-11 1 ## Usp24 5.126661e-16 0.6640007 0.686 0.370 2.045794e-11 1 ## Gm46528 5.183093e-16 1.0651357 0.143 0.027 2.068313e-11 1 ## Rundc3b 5.427563e-16 0.5667104 0.657 0.328 2.165869e-11 1 ## Tecpr11 5.711122e-16 0.4078342 0.886 0.542 2.279023e-11 1 ## Mag 5.835410e-16 1.3995879 0.224 0.062 2.328620e-11 1 ## Paip21 6.050925e-16 0.4967684 0.967 0.718 2.414621e-11 1 ## Adh51 6.581227e-16 0.6013856 0.845 0.503 2.626239e-11 1 ## Smim11 7.479172e-16 0.6055793 0.678 0.345 2.984564e-11 1 ## Eid11 7.807051e-16 0.3549858 0.992 0.739 3.115404e-11 1 ## Mrpl521 7.808792e-16 0.4817570 0.976 0.703 3.116099e-11 1 ## Fam89a 8.665364e-16 1.9143899 0.139 0.026 3.457913e-11 1 ## Dctn6 8.805865e-16 0.5981550 0.731 0.428 3.513981e-11 1 ## Ubl51 8.849994e-16 0.4080281 1.000 0.787 3.531590e-11 1 ## Muc13 8.892922e-16 0.2772540 1.000 0.899 3.548720e-11 1 ## Glra3 9.056539e-16 5.1275362 0.045 0.000 3.614012e-11 1 ## Snhg4 9.100162e-16 0.8081217 0.478 0.216 3.631420e-11 1 ## Naca 9.554498e-16 0.1826438 0.996 0.812 3.812722e-11 1 ## Sytl2 9.758651e-16 1.0982112 0.208 0.057 3.894189e-11 1 ## Zfp709 1.011614e-15 1.6288226 0.180 0.043 4.036845e-11 1 ## Kcnh61 1.055048e-15 0.2987322 0.963 0.600 4.210170e-11 1 ## Ift25 1.059929e-15 0.7971504 0.514 0.250 4.229647e-11 1 ## Mapre2 1.080374e-15 0.7927441 0.510 0.240 4.311234e-11 1 ## Zfp961 1.104921e-15 1.1808402 0.298 0.106 4.409188e-11 1 ## Rpl23a 1.118514e-15 0.1584530 0.959 0.661 4.463428e-11 1 ## Gusb1 1.145146e-15 0.7479510 0.584 0.291 4.569706e-11 1 ## Hnrnpa0 1.203163e-15 0.4289081 0.943 0.655 4.801221e-11 1 ## Pdcd7 1.215127e-15 0.5427130 0.633 0.336 4.848965e-11 1 ## Septin21 1.235273e-15 0.5437663 0.902 0.608 4.929355e-11 1 ## Ptov11 1.246776e-15 0.5010426 0.976 0.709 4.975260e-11 1 ## Inpp5f 1.263839e-15 1.2651923 0.306 0.109 5.043350e-11 1 ## Prorsd1 1.265094e-15 0.5660981 0.616 0.310 5.048356e-11 1 ## Emc71 1.272143e-15 0.6084676 0.865 0.583 5.076485e-11 1 ## Rhoa1 1.393590e-15 0.3626544 0.996 0.849 5.561120e-11 1 ## Rraga1 1.599911e-15 0.6217325 0.792 0.494 6.384444e-11 1 ## Klhdc21 1.629775e-15 0.6655719 0.808 0.513 6.503619e-11 1 ## Pik3c2b 1.641829e-15 0.6821172 0.404 0.169 6.551717e-11 1 ## Gm51641 1.688838e-15 3.4066217 0.061 0.003 6.739310e-11 1 ## Cntnap5b 1.780464e-15 5.1177534 0.057 0.002 7.104941e-11 1 ## Orm1 1.792935e-15 4.6551072 0.049 0.001 7.154708e-11 1 ## Cyp4a32 1.827968e-15 1.8398762 0.127 0.022 7.294506e-11 1 ## Dynlrb11 1.845219e-15 0.4246217 0.955 0.689 7.363345e-11 1 ## Cryba21 1.957268e-15 0.5198441 0.816 0.472 7.810479e-11 1 ## Spg21 2.022372e-15 0.7104925 0.645 0.342 8.070276e-11 1 ## Anp32e 2.081506e-15 0.5014188 0.714 0.381 8.306250e-11 1 ## Ccl27a 2.084968e-15 1.1677064 0.310 0.115 8.320064e-11 1 ## Snrpf 2.121662e-15 0.1710871 0.898 0.589 8.466491e-11 1 ## Krt4 2.187100e-15 4.2270689 0.053 0.001 8.727624e-11 1 ## Erp291 2.371968e-15 0.4961703 0.935 0.686 9.465339e-11 1 ## Sugt11 2.387454e-15 0.4933929 0.849 0.530 9.527135e-11 1 ## Slc37a3 2.442823e-15 0.7034560 0.543 0.275 9.748085e-11 1 ## Pofut2 2.473254e-15 0.8320180 0.629 0.334 9.869519e-11 1 ## C230034O21Rik 2.507054e-15 2.6351583 0.094 0.011 1.000440e-10 1 ## Dcxr 2.515709e-15 0.6407736 0.620 0.321 1.003894e-10 1 ## Mrps18c1 2.640378e-15 0.5754873 0.812 0.486 1.053643e-10 1 ## Fahd1 2.685963e-15 0.3135810 0.624 0.314 1.071833e-10 1 ## Izumo4 2.809547e-15 0.6410314 0.482 0.217 1.121150e-10 1 ## Pin1 2.858840e-15 0.4674202 0.751 0.428 1.140820e-10 1 ## Gm15706 2.885552e-15 1.7836643 0.237 0.075 1.151480e-10 1 ## Cd91 2.889269e-15 0.7553260 0.792 0.490 1.152963e-10 1 ## Fahd2a 2.983262e-15 0.8492837 0.388 0.164 1.190471e-10 1 ## A630066F11Rik 3.021815e-15 1.6330003 0.196 0.052 1.205855e-10 1 ## Cirbp 3.062282e-15 0.8140783 0.727 0.455 1.222004e-10 1 ## Rpl26 3.128455e-15 0.2208425 1.000 0.891 1.248410e-10 1 ## Rpa3 3.252765e-15 0.6746536 0.510 0.235 1.298016e-10 1 ## Stmp1 3.259490e-15 0.4583435 0.914 0.577 1.300699e-10 1 ## Fcna 3.314676e-15 1.2658007 0.265 0.087 1.322721e-10 1 ## Foxo1 3.364742e-15 0.6362852 0.567 0.293 1.342700e-10 1 ## Sdhaf4 3.539830e-15 0.7434114 0.714 0.417 1.412569e-10 1 ## Gm38487 3.571481e-15 1.1558101 0.188 0.048 1.425199e-10 1 ## Dusp23 3.638083e-15 0.9226120 0.380 0.154 1.451777e-10 1 ## Ppp1r9a 3.747489e-15 0.5144030 0.567 0.281 1.495435e-10 1 ## Clic11 3.884242e-15 0.3561692 0.996 0.784 1.550007e-10 1 ## Far1 3.885598e-15 0.6870949 0.384 0.151 1.550548e-10 1 ## Armcx4 4.275882e-15 2.0254353 0.135 0.026 1.706291e-10 1 ## Rnf111 4.369290e-15 0.5096849 0.812 0.490 1.743565e-10 1 ## Tprg1l1 4.377568e-15 0.5878295 0.816 0.509 1.746868e-10 1 ## Smim10l11 4.558911e-15 0.4704910 0.894 0.621 1.819234e-10 1 ## Erlec1 4.588469e-15 0.5009916 0.857 0.586 1.831029e-10 1 ## Vsig1 4.645555e-15 3.4249345 0.078 0.007 1.853809e-10 1 ## Czib 4.703857e-15 0.6745721 0.580 0.302 1.877074e-10 1 ## Coq7 5.128459e-15 0.6566804 0.612 0.305 2.046511e-10 1 ## Lamc1 5.256551e-15 0.8474734 0.396 0.165 2.097627e-10 1 ## Nefm 5.324091e-15 0.5299320 0.380 0.152 2.124579e-10 1 ## Spock2 5.568160e-15 0.5794677 0.645 0.324 2.221974e-10 1 ## Ccdc921 5.834303e-15 0.5472854 0.608 0.302 2.328178e-10 1 ## Abhd14a 6.083551e-15 0.8555387 0.404 0.171 2.427641e-10 1 ## Tigd2 6.183938e-15 0.5416669 0.604 0.298 2.467700e-10 1 ## Rtl8b 6.249814e-15 0.9373799 0.343 0.129 2.493988e-10 1 ## Emb 6.323254e-15 0.6722990 0.441 0.189 2.523294e-10 1 ## Lipt2 6.514713e-15 1.7611507 0.188 0.052 2.599696e-10 1 ## Gstm5 6.582259e-15 0.6298972 0.637 0.350 2.626650e-10 1 ## Apool 6.609025e-15 0.7415996 0.429 0.196 2.637331e-10 1 ## Clic6 6.679947e-15 1.2534901 0.249 0.082 2.665633e-10 1 ## Naxd1 6.790855e-15 0.4404110 0.894 0.549 2.709891e-10 1 ## Cimap1b 6.964192e-15 1.0023665 0.371 0.152 2.779061e-10 1 ## Ucp21 7.056423e-15 0.3336678 0.996 0.850 2.815866e-10 1 ## Ufc11 7.126944e-15 0.5393128 0.927 0.653 2.844007e-10 1 ## Tbpl1 7.139903e-15 0.6711962 0.527 0.257 2.849178e-10 1 ## Mtln 7.426771e-15 0.6341926 0.665 0.353 2.963653e-10 1 ## Psmb8 7.821835e-15 0.3545630 0.931 0.590 3.121303e-10 1 ## Morf4l11 7.935062e-15 0.5513777 0.902 0.628 3.166486e-10 1 ## Prnp1 7.999322e-15 0.3379827 1.000 0.826 3.192130e-10 1 ## Dnajc4 8.117033e-15 0.7705147 0.457 0.212 3.239102e-10 1 ## Axdnd1 8.384888e-15 3.0918271 0.065 0.004 3.345989e-10 1 ## As3mt 8.783487e-15 0.6841276 0.710 0.400 3.505050e-10 1 ## Aopep 8.923955e-15 0.3684969 0.943 0.650 3.561104e-10 1 ## Dnajc19 8.974814e-15 0.3855640 0.845 0.497 3.581399e-10 1 ## Nek7 9.769556e-15 0.5772545 0.514 0.248 3.898541e-10 1 ## Mrps16 9.976587e-15 0.4514719 0.829 0.491 3.981157e-10 1 ## Capn2 9.993078e-15 0.8995263 0.339 0.129 3.987738e-10 1 ## Gm12146 1.044890e-14 1.0638034 0.257 0.085 4.169635e-10 1 ## Norad1 1.048687e-14 0.4914367 0.918 0.634 4.184786e-10 1 ## Pet1001 1.059531e-14 0.4785343 0.918 0.614 4.228060e-10 1 ## Spr 1.109867e-14 0.4516256 0.702 0.397 4.428925e-10 1 ## Ctbs 1.124558e-14 1.1650422 0.233 0.073 4.487548e-10 1 ## 6330407A03Rik 1.176768e-14 0.7093517 0.253 0.081 4.695891e-10 1 ## Tox1 1.190621e-14 0.5299522 0.792 0.484 4.751173e-10 1 ## Lrrc58 1.222699e-14 0.5361515 0.882 0.589 4.879180e-10 1 ## Tmem230 1.316654e-14 0.7112027 0.506 0.247 5.254106e-10 1 ## Psmg41 1.318355e-14 0.5269485 0.669 0.352 5.260896e-10 1 ## Ints6l 1.336598e-14 0.4266048 0.359 0.140 5.333695e-10 1 ## Zdhhc141 1.339971e-14 0.4718501 0.604 0.295 5.347155e-10 1 ## Snx6 1.343503e-14 0.4869390 0.853 0.544 5.361250e-10 1 ## Gm53896 1.362391e-14 1.7395501 0.159 0.037 5.436621e-10 1 ## Sdf2l1 1.423585e-14 0.6572598 0.678 0.379 5.680817e-10 1 ## Rab9 1.425332e-14 0.5442532 0.604 0.323 5.687789e-10 1 ## Dcaf8 1.432037e-14 0.4435623 0.804 0.479 5.714544e-10 1 ## Trp53i111 1.464510e-14 0.4447788 0.922 0.581 5.844128e-10 1 ## Tmem106c 1.473825e-14 1.0002493 0.437 0.197 5.881298e-10 1 ## Psmb101 1.482306e-14 0.3222314 0.955 0.692 5.915141e-10 1 ## Gas21 1.540113e-14 1.1534177 0.363 0.150 6.145819e-10 1 ## Aimp11 1.544285e-14 0.5079494 0.935 0.648 6.162469e-10 1 ## Ankrd13d 1.599818e-14 2.1341177 0.102 0.015 6.384072e-10 1 ## Cdkl2 1.606554e-14 0.8543519 0.351 0.138 6.410955e-10 1 ## Slc25a36 1.670185e-14 0.4911982 0.727 0.426 6.664872e-10 1 ## Tmem150b 1.677301e-14 1.2636453 0.220 0.069 6.693269e-10 1 ## Rps27 1.694017e-14 0.2275434 0.984 0.849 6.759975e-10 1 ## Arl6 1.698568e-14 1.1773745 0.294 0.106 6.778137e-10 1 ## Map3k151 1.706795e-14 0.6870291 0.449 0.198 6.810964e-10 1 ## Med21 1.725981e-14 0.8686704 0.510 0.256 6.887529e-10 1 ## Xpnpep21 1.776507e-14 0.4458126 0.751 0.416 7.089150e-10 1 ## Colgalt2 1.822660e-14 5.0227823 0.041 0.000 7.273325e-10 1 ## Gm7361 1.822660e-14 5.8118242 0.041 0.000 7.273325e-10 1 ## Psme11 1.915313e-14 0.4119439 0.971 0.731 7.643058e-10 1 ## Ctss 1.924279e-14 0.6019407 0.404 0.176 7.678836e-10 1 ## Jpx 1.982160e-14 1.6941107 0.180 0.047 7.909808e-10 1 ## Nudcd2 1.990488e-14 0.6684650 0.571 0.285 7.943041e-10 1 ## Porcn 2.019065e-14 1.6715399 0.139 0.029 8.057077e-10 1 ## Rcn2 2.083094e-14 0.5769027 0.629 0.333 8.312586e-10 1 ## Hypk1 2.085349e-14 0.4422325 0.951 0.644 8.321583e-10 1 ## Gnai21 2.088179e-14 0.3595143 0.988 0.818 8.332880e-10 1 ## Dmd 2.221178e-14 0.4292839 0.469 0.217 8.863609e-10 1 ## Lamtor41 2.316271e-14 0.4982399 0.947 0.623 9.243078e-10 1 ## Trappc14 2.389347e-14 0.5430023 0.727 0.434 9.534690e-10 1 ## Ostf1 2.412139e-14 0.3712166 0.763 0.452 9.625642e-10 1 ## Rnf135 2.469981e-14 1.4310591 0.233 0.073 9.856460e-10 1 ## Cbfb 2.557787e-14 0.5951388 0.694 0.394 1.020685e-09 1 ## Cct5 2.571509e-14 0.4644226 0.951 0.658 1.026161e-09 1 ## Cisd11 2.580151e-14 0.4104230 0.951 0.648 1.029609e-09 1 ## Cd1d1 2.659689e-14 1.3604984 0.224 0.071 1.061349e-09 1 ## Bcas2 2.854423e-14 0.4088794 0.841 0.509 1.139057e-09 1 ## Meis2 2.888176e-14 1.0680464 0.245 0.078 1.152526e-09 1 ## Ei241 3.246769e-14 0.5247046 0.804 0.507 1.295623e-09 1 ## Tap2 3.299967e-14 0.8217734 0.445 0.203 1.316852e-09 1 ## Sdhd 3.438411e-14 0.2505999 0.882 0.570 1.372098e-09 1 ## Homer1 3.460071e-14 0.6198275 0.400 0.177 1.380741e-09 1 ## Lyn 3.612760e-14 0.6298613 0.580 0.323 1.441672e-09 1 ## Tbcb1 3.701327e-14 0.4164163 0.910 0.643 1.477015e-09 1 ## Rpp25l 3.739924e-14 0.6800820 0.571 0.302 1.492417e-09 1 ## Med281 3.770501e-14 0.4386080 0.914 0.611 1.504619e-09 1 ## Arf4 3.820736e-14 0.3912304 0.935 0.648 1.524665e-09 1 ## Lsm8 3.838081e-14 0.3886519 0.641 0.340 1.531586e-09 1 ## Pvt1 3.842533e-14 3.0806035 0.053 0.002 1.533363e-09 1 ## Ppp1r111 4.023082e-14 0.4932872 0.873 0.594 1.605411e-09 1 ## Ephx4 4.045384e-14 1.1428108 0.192 0.052 1.614311e-09 1 ## Sdcbp2 4.116657e-14 0.3437085 0.898 0.579 1.642752e-09 1 ## Smad11 4.184716e-14 0.4758732 0.918 0.652 1.669911e-09 1 ## Sun3 4.228572e-14 1.1852463 0.184 0.049 1.687412e-09 1 ## Vdac2 4.237207e-14 0.2440060 0.992 0.759 1.690857e-09 1 ## Rad51d 4.292127e-14 0.6201890 0.449 0.204 1.712773e-09 1 ## Fibp 4.361280e-14 0.6266116 0.727 0.442 1.740369e-09 1 ## Lrp3 4.363203e-14 0.7372076 0.392 0.161 1.741136e-09 1 ## Psmb11 4.412375e-14 0.4077701 0.984 0.758 1.760758e-09 1 ## Atp8a11 4.720669e-14 0.2959059 0.808 0.442 1.883783e-09 1 ## Zbp1 4.757479e-14 0.7240055 0.355 0.143 1.898472e-09 1 ## Mid2 4.961273e-14 0.9426217 0.306 0.116 1.979796e-09 1 ## Micos13 5.237486e-14 0.4254485 0.984 0.753 2.090019e-09 1 ## Tmed101 5.316850e-14 0.4558798 0.939 0.702 2.121689e-09 1 ## Eif11 5.442384e-14 0.3417608 1.000 0.908 2.171783e-09 1 ## Scamp1 5.496717e-14 0.5210279 0.776 0.479 2.193465e-09 1 ## Cda 5.662363e-14 0.2138957 0.918 0.637 2.259566e-09 1 ## Fundc2 5.700398e-14 0.4066328 0.665 0.347 2.274744e-09 1 ## Eif2s21 5.736199e-14 0.4041826 0.980 0.781 2.289030e-09 1 ## Arid5b 5.761637e-14 0.4852679 0.686 0.396 2.299181e-09 1 ## Zc3h12c 5.835715e-14 2.1441919 0.122 0.023 2.328742e-09 1 ## Hint2 6.412051e-14 0.4688278 0.776 0.447 2.558729e-09 1 ## Prodh21 6.576469e-14 0.2884019 0.722 0.379 2.624340e-09 1 ## Psma21 6.697726e-14 0.3976162 0.971 0.692 2.672728e-09 1 ## B9d1 6.761795e-14 0.6941329 0.559 0.283 2.698294e-09 1 ## Gm26871 6.911498e-14 0.2040499 0.857 0.511 2.758033e-09 1 ## Mettl23 7.271617e-14 0.6188538 0.584 0.303 2.901739e-09 1 ## Atp5pf 8.151641e-14 0.2077607 1.000 0.842 3.252912e-09 1 ## Eif3i 8.337580e-14 0.3291986 0.939 0.634 3.327111e-09 1 ## Ubb1 8.817865e-14 0.3106281 1.000 0.965 3.518769e-09 1 ## Bcl2 8.961444e-14 1.5749490 0.167 0.044 3.576064e-09 1 ## Rpp211 9.466526e-14 0.5261873 0.771 0.469 3.777617e-09 1 ## Bnip3l1 9.883278e-14 0.4467460 0.886 0.563 3.943922e-09 1 ## Angptl6 1.010422e-13 1.5661590 0.200 0.059 4.032090e-09 1 ## Ndufa8 1.012504e-13 0.3737066 0.939 0.625 4.040398e-09 1 ## Foxa31 1.040551e-13 0.4148317 0.759 0.421 4.152320e-09 1 ## S100a131 1.066899e-13 0.3235745 0.992 0.746 4.257461e-09 1 ## Glrx3 1.078434e-13 0.4558024 0.837 0.502 4.303492e-09 1 ## Cog11 1.114943e-13 0.4481686 0.682 0.373 4.449180e-09 1 ## Gbp7 1.159085e-13 0.9740744 0.257 0.088 4.625328e-09 1 ## Dgcr6 1.205225e-13 0.5022893 0.755 0.454 4.809449e-09 1 ## Gm15972 1.211479e-13 2.6642948 0.122 0.024 4.834406e-09 1 ## Csrp1 1.222148e-13 0.9511946 0.335 0.140 4.876980e-09 1 ## Jkamp1 1.248245e-13 0.3820315 0.706 0.401 4.981120e-09 1 ## Churc11 1.249734e-13 0.4947115 0.845 0.519 4.987065e-09 1 ## Ndufaf3 1.288927e-13 0.6983688 0.637 0.365 5.143465e-09 1 ## Vamp31 1.327155e-13 0.5209746 0.808 0.530 5.296013e-09 1 ## Hdac7 1.394199e-13 0.5869285 0.392 0.168 5.563551e-09 1 ## Ppargc1b 1.399815e-13 0.5179095 0.380 0.162 5.585961e-09 1 ## Nsf1 1.404429e-13 0.4459300 0.718 0.416 5.604374e-09 1 ## Cryl1 1.564741e-13 0.4697070 0.502 0.242 6.244101e-09 1 ## Gm10400 1.634130e-13 0.7287177 0.184 0.052 6.520996e-09 1 ## Katnal1 1.671323e-13 1.1012798 0.249 0.086 6.669415e-09 1 ## Tmem42 1.796883e-13 0.6248001 0.571 0.299 7.170463e-09 1 ## Sidt1 1.854744e-13 0.7889278 0.351 0.143 7.401356e-09 1 ## Mri1 1.882283e-13 0.8807169 0.363 0.159 7.511252e-09 1 ## Ccdc126 1.936976e-13 1.2250763 0.229 0.073 7.729505e-09 1 ## Cox6a11 1.977524e-13 0.2556486 0.988 0.778 7.891311e-09 1 ## Nme31 2.047144e-13 0.6578678 0.751 0.477 8.169126e-09 1 ## Ciao2b1 2.063201e-13 0.4494128 0.763 0.454 8.233203e-09 1 ## Gabbr2 2.097676e-13 2.4795162 0.082 0.010 8.370777e-09 1 ## Akr1c131 2.107978e-13 0.3911652 0.971 0.657 8.411884e-09 1 ## Tubb2a1 2.113664e-13 0.5648034 0.555 0.282 8.434575e-09 1 ## Lin37 2.130351e-13 0.6306128 0.506 0.255 8.501165e-09 1 ## D130020L05Rik 2.182981e-13 1.1272770 0.233 0.078 8.711185e-09 1 ## Hsd17b7 2.194775e-13 0.9500591 0.412 0.194 8.758248e-09 1 ## Gm28523 2.235366e-13 1.1767167 0.171 0.045 8.920227e-09 1 ## Gars1 2.244685e-13 0.3553401 0.927 0.628 8.957417e-09 1 ## Gm32546 2.309298e-13 2.5204151 0.082 0.010 9.215255e-09 1 ## Pgap2 2.332793e-13 0.6756508 0.465 0.226 9.309010e-09 1 ## Atp8b5 2.433590e-13 1.2692546 0.155 0.038 9.711240e-09 1 ## Tor3a1 2.450140e-13 0.5697107 0.620 0.332 9.777283e-09 1 ## Naaa 2.493348e-13 0.7483198 0.424 0.199 9.949707e-09 1 ## Lpcat2 2.517248e-13 2.6109274 0.057 0.003 1.004508e-08 1 ## Mbnl21 2.762456e-13 0.3944497 0.959 0.745 1.102358e-08 1 ## Gm39369 2.826435e-13 1.2798984 0.180 0.050 1.127889e-08 1 ## Rps6ka3 2.937872e-13 0.4011492 0.763 0.458 1.172358e-08 1 ## Rnf71 2.988350e-13 0.5046489 0.820 0.532 1.192501e-08 1 ## Zfp771 3.113824e-13 0.7651766 0.514 0.262 1.242571e-08 1 ## Ccdc30 3.114773e-13 0.9962728 0.159 0.040 1.242950e-08 1 ## Ift1721 3.128426e-13 0.6950496 0.686 0.405 1.248398e-08 1 ## Gimap5 3.136336e-13 0.7668817 0.310 0.121 1.251555e-08 1 ## Zfp422 3.139257e-13 0.5665788 0.510 0.249 1.252721e-08 1 ## Gm12576 3.147331e-13 0.8020211 0.380 0.169 1.255943e-08 1 ## Tmem1471 3.184917e-13 0.4642404 0.845 0.553 1.270941e-08 1 ## Nans 3.255076e-13 0.4469374 0.596 0.308 1.298938e-08 1 ## Oaz11 3.284476e-13 0.3640124 0.996 0.876 1.310670e-08 1 ## Snap471 3.349800e-13 0.7038842 0.551 0.284 1.336738e-08 1 ## Samhd1 3.567237e-13 0.5759018 0.616 0.333 1.423506e-08 1 ## 4921528I07Rik 3.672549e-13 4.7096883 0.037 0.000 1.465531e-08 1 ## Mcrip11 3.679337e-13 0.5148571 0.767 0.450 1.468239e-08 1 ## Emc9 3.703500e-13 1.1585705 0.343 0.147 1.477882e-08 1 ## Cotl11 3.751957e-13 0.3170241 0.959 0.660 1.497218e-08 1 ## C230035I16Rik 3.756306e-13 3.1490068 0.102 0.017 1.498954e-08 1 ## Cebpa 3.816864e-13 0.8483127 0.408 0.188 1.523119e-08 1 ## Pop7 3.858356e-13 0.8202807 0.551 0.307 1.539677e-08 1 ## Trappc31 3.864670e-13 0.6523260 0.722 0.433 1.542197e-08 1 ## Ttll5 3.865632e-13 0.4366501 0.441 0.201 1.542580e-08 1 ## Marchf2 3.963520e-13 0.6467495 0.527 0.274 1.581643e-08 1 ## Swi5 3.965586e-13 0.3907637 0.951 0.706 1.582467e-08 1 ## Glb11 4.098590e-13 0.6882007 0.576 0.305 1.635542e-08 1 ## Ipo8 4.150756e-13 0.4833094 0.600 0.306 1.656359e-08 1 ## Fbxo211 4.240895e-13 0.5318392 0.608 0.311 1.692329e-08 1 ## Nt5e1 4.517974e-13 0.1649344 0.971 0.604 1.802898e-08 1 ## Eef1akmt1 4.626841e-13 0.8003106 0.514 0.266 1.846341e-08 1 ## Armcx1 4.663271e-13 0.4479008 0.531 0.260 1.860878e-08 1 ## Cct31 4.771873e-13 0.4462294 0.890 0.568 1.904216e-08 1 ## Tmbim4 4.977391e-13 0.4477792 0.914 0.600 1.986228e-08 1 ## Dpy30 5.096066e-13 0.5047478 0.861 0.540 2.033585e-08 1 ## Pdcd51 5.170616e-13 0.4490093 0.927 0.644 2.063334e-08 1 ## Mrpl531 5.199364e-13 0.4156240 0.869 0.590 2.074806e-08 1 ## Cfdp1 5.282326e-13 0.5018590 0.808 0.502 2.107912e-08 1 ## Parp14 5.440667e-13 0.2215507 0.600 0.317 2.171098e-08 1 ## Tnks1bp1 5.554886e-13 0.4331663 0.653 0.359 2.216677e-08 1 ## Svbp 5.664015e-13 0.8687478 0.339 0.145 2.260225e-08 1 ## Tvp23b1 5.816314e-13 0.4857182 0.820 0.523 2.321000e-08 1 ## Atl3 6.018176e-13 0.9098207 0.351 0.150 2.401553e-08 1 ## Il3ra 6.023596e-13 0.6313513 0.331 0.132 2.403716e-08 1 ## Fbxw17 6.072549e-13 0.9608441 0.294 0.110 2.423251e-08 1 ## Ror2 6.138826e-13 0.9927725 0.282 0.108 2.449698e-08 1 ## Gnaq 6.182347e-13 0.4102483 0.845 0.552 2.467066e-08 1 ## Rbpms 6.194074e-13 0.7999538 0.420 0.187 2.471745e-08 1 ## Ccdc86 6.247519e-13 0.6779525 0.404 0.181 2.493073e-08 1 ## Ccpg1 6.354480e-13 0.5147135 0.792 0.472 2.535755e-08 1 ## Rras1 6.485940e-13 0.8047904 0.539 0.287 2.588214e-08 1 ## Pgbd5 6.552049e-13 1.3163113 0.171 0.046 2.614595e-08 1 ## Mrpl571 6.566897e-13 0.4330933 0.902 0.590 2.620520e-08 1 ## Fgf18 6.680077e-13 3.3625597 0.049 0.002 2.665685e-08 1 ## Scin 6.717873e-13 1.5971466 0.347 0.157 2.680767e-08 1 ## Smap2 6.854690e-13 0.6012248 0.637 0.370 2.735364e-08 1 ## Pyurf 6.882271e-13 0.6114762 0.612 0.342 2.746370e-08 1 ## Macroh2a2 7.056052e-13 1.0735047 0.261 0.093 2.815718e-08 1 ## Casd1 7.194276e-13 0.6645583 0.567 0.314 2.870876e-08 1 ## Csgalnact2 7.437204e-13 0.8791170 0.220 0.071 2.967816e-08 1 ## Ccdc28b 7.475512e-13 0.7302925 0.469 0.231 2.983103e-08 1 ## Nrp2 7.687037e-13 0.9235162 0.163 0.043 3.067512e-08 1 ## Rab25 7.843877e-13 0.3670222 0.927 0.651 3.130099e-08 1 ## Mrpl43 7.847131e-13 0.4696105 0.857 0.535 3.131398e-08 1 ## Fam216a 7.929353e-13 0.6538088 0.486 0.238 3.164208e-08 1 ## 1600014C10Rik 8.108305e-13 0.3509768 0.420 0.192 3.235619e-08 1 ## Gm41603 8.250210e-13 2.4157688 0.102 0.017 3.292246e-08 1 ## Sap18 8.652681e-13 0.4242556 0.845 0.539 3.452852e-08 1 ## Ilk 8.751627e-13 0.5074385 0.567 0.309 3.492337e-08 1 ## Msantd3 8.949989e-13 1.6538325 0.151 0.038 3.571493e-08 1 ## C1galt1c1 8.984061e-13 0.4781030 0.637 0.375 3.585089e-08 1 ## Fam221a 9.063265e-13 0.5532458 0.584 0.309 3.616696e-08 1 ## Pde2a1 9.237281e-13 0.4734813 0.649 0.356 3.686137e-08 1 ## Rnf114 9.261490e-13 0.4221611 0.637 0.358 3.695798e-08 1 ## Eif5a 9.595262e-13 0.2660608 0.996 0.796 3.828989e-08 1 ## Tex30 9.812995e-13 0.7770410 0.388 0.177 3.915876e-08 1 ## Tmem242 9.896848e-13 0.6404953 0.429 0.208 3.949337e-08 1 ## Lsm11 9.920296e-13 1.2879614 0.224 0.077 3.958694e-08 1 ## Unc79 1.000105e-12 0.2868786 0.653 0.342 3.990918e-08 1 ## Cct71 1.009212e-12 0.4040972 0.971 0.716 4.027259e-08 1 ## Polr2d 1.033104e-12 0.9416682 0.359 0.157 4.122603e-08 1 ## Mindy21 1.059162e-12 0.3234452 0.894 0.588 4.226587e-08 1 ## Rhob1 1.066304e-12 0.3852096 0.869 0.574 4.255084e-08 1 ## Slc12a5 1.087376e-12 1.9946652 0.086 0.012 4.339175e-08 1 ## Wdfy2 1.092777e-12 0.8563873 0.294 0.116 4.360726e-08 1 ## Ttc21b 1.111984e-12 1.1811086 0.253 0.090 4.437371e-08 1 ## Ywhag1 1.125427e-12 0.4521390 0.796 0.494 4.491017e-08 1 ## Cox6b11 1.128439e-12 0.2363459 0.996 0.829 4.503036e-08 1 ## Acsf3 1.178913e-12 0.8114728 0.416 0.199 4.704452e-08 1 ## Ccdc107 1.190951e-12 0.4052500 0.735 0.407 4.752490e-08 1 ## Cnot6l1 1.203118e-12 0.3799443 0.971 0.767 4.801043e-08 1 ## Chchd11 1.206183e-12 0.3632674 0.857 0.539 4.813274e-08 1 ## Sec61a11 1.236488e-12 0.3796822 0.927 0.626 4.934204e-08 1 ## Ube2b 1.249481e-12 0.3166075 0.988 0.772 4.986053e-08 1 ## Itgb1bp1 1.320795e-12 0.8639514 0.351 0.150 5.270633e-08 1 ## Polr2j1 1.353043e-12 0.4747508 0.857 0.568 5.399318e-08 1 ## Fmc1 1.395428e-12 0.4286138 0.706 0.403 5.568456e-08 1 ## Gm16174 1.406234e-12 1.4197677 0.155 0.039 5.611576e-08 1 ## Lamtor5 1.492305e-12 0.4116087 0.759 0.456 5.955045e-08 1 ## Nme11 1.499509e-12 0.2582610 0.931 0.632 5.983790e-08 1 ## Eloc1 1.508143e-12 0.4832565 0.914 0.610 6.018244e-08 1 ## Rai2 1.511714e-12 0.5521150 0.482 0.233 6.032497e-08 1 ## 4930594M22Rik 1.527823e-12 2.9156103 0.073 0.008 6.096777e-08 1 ## Ndufa41 1.531802e-12 0.2593390 0.992 0.808 6.112657e-08 1 ## Hddc3 1.602495e-12 1.4699816 0.155 0.041 6.394755e-08 1 ## Ndufb5 1.655747e-12 0.2796291 0.943 0.697 6.607259e-08 1 ## Ilkap1 1.714274e-12 0.4382346 0.816 0.486 6.840810e-08 1 ## Ap2b11 1.719013e-12 0.4003343 0.918 0.641 6.859723e-08 1 ## Tmie 1.756850e-12 0.6949854 0.404 0.182 7.010709e-08 1 ## Uqcrh 1.765110e-12 0.2149954 1.000 0.826 7.043671e-08 1 ## Gnaz1 1.767008e-12 0.3587997 0.637 0.346 7.051244e-08 1 ## Bace11 1.821055e-12 0.4952294 0.829 0.535 7.266920e-08 1 ## Arhgap6 1.825519e-12 1.7475974 0.098 0.017 7.284734e-08 1 ## Trir1 1.876437e-12 0.3844626 0.967 0.713 7.487923e-08 1 ## Cmc1 1.880051e-12 0.5591443 0.624 0.340 7.502345e-08 1 ## Borcs6 1.965317e-12 0.7193788 0.408 0.182 7.842597e-08 1 ## Gm34294 2.004378e-12 3.0116785 0.057 0.004 7.998472e-08 1 ## Phax 2.064499e-12 0.5598230 0.653 0.389 8.238385e-08 1 ## Uap1 2.079327e-12 0.6421135 0.482 0.245 8.297555e-08 1 ## Rab3a1 2.153410e-12 0.6102643 0.498 0.240 8.593183e-08 1 ## Xaf1 2.244044e-12 0.8834161 0.257 0.094 8.954859e-08 1 ## Trp53bp1 2.262400e-12 0.3064393 0.629 0.342 9.028109e-08 1 ## Paip2b 2.306055e-12 0.7480072 0.412 0.189 9.202312e-08 1 ## Gm10029 2.312566e-12 0.2868259 0.841 0.506 9.228293e-08 1 ## Cdc421 2.346923e-12 0.2963014 0.992 0.851 9.365396e-08 1 ## Psmg2 2.355622e-12 0.5040299 0.437 0.205 9.400108e-08 1 ## Ptrhd1 2.373365e-12 0.7791364 0.461 0.232 9.470912e-08 1 ## Baiap2l1 2.423989e-12 0.2863414 0.600 0.333 9.672929e-08 1 ## Phb1 2.426419e-12 0.2247710 0.833 0.527 9.682625e-08 1 ## Arl1 2.469768e-12 0.3685710 0.914 0.629 9.855610e-08 1 ## Tnr 2.500368e-12 0.5969830 0.298 0.117 9.977717e-08 1 ## Dhrs7 2.538055e-12 0.6567330 0.604 0.328 1.012811e-07 1 ## Abcc41 2.600280e-12 0.8368200 0.327 0.133 1.037642e-07 1 ## Mtmr71 2.610438e-12 0.4817021 0.694 0.410 1.041695e-07 1 ## Dmxl2 2.828421e-12 0.7368203 0.245 0.085 1.128681e-07 1 ## Cox5b 2.843134e-12 0.1604726 0.992 0.800 1.134552e-07 1 ## Selenom1 2.845642e-12 0.3725024 1.000 0.832 1.135554e-07 1 ## Glmn 2.925973e-12 1.2751288 0.237 0.087 1.167610e-07 1 ## 1700066M21Rik 2.950303e-12 1.3951052 0.151 0.040 1.177318e-07 1 ## Adgrg7 3.169469e-12 0.4782085 0.510 0.258 1.264777e-07 1 ## Tmem1651 3.177069e-12 0.6284622 0.629 0.350 1.267809e-07 1 ## Nop10 3.214265e-12 0.2426025 0.820 0.486 1.282652e-07 1 ## Smim30 3.273652e-12 0.6159923 0.494 0.269 1.306351e-07 1 ## Nap1l5 3.309570e-12 1.4927018 0.167 0.048 1.320684e-07 1 ## Lpar5 3.331541e-12 1.0087383 0.343 0.157 1.329452e-07 1 ## Kbtbd12 3.342267e-12 3.8050824 0.053 0.003 1.333732e-07 1 ## Ddost 3.370970e-12 0.3976047 0.939 0.635 1.345186e-07 1 ## D830035M03Rik 3.399718e-12 3.8110601 0.053 0.003 1.356658e-07 1 ## 4933412E12Rik 3.507929e-12 1.8965870 0.110 0.022 1.399839e-07 1 ## Slc25a29 3.554880e-12 1.0501812 0.220 0.075 1.418575e-07 1 ## Lamp21 3.614683e-12 0.4529736 0.918 0.709 1.442439e-07 1 ## Spata45 3.701817e-12 3.2935888 0.053 0.003 1.477210e-07 1 ## 2310039H08Rik 3.713574e-12 0.5089720 0.710 0.411 1.481902e-07 1 ## Ctsz 3.777783e-12 0.3317272 0.959 0.641 1.507524e-07 1 ## Yy11 3.847260e-12 0.4173127 0.922 0.699 1.535249e-07 1 ## Erp44 3.859310e-12 0.4272371 0.731 0.449 1.540058e-07 1 ## Lhfpl4 3.942393e-12 1.1868731 0.171 0.048 1.573212e-07 1 ## Scmh1 3.958792e-12 0.4796678 0.555 0.292 1.579756e-07 1 ## Tmem151b 3.960043e-12 1.0474164 0.220 0.075 1.580255e-07 1 ## Impdh11 4.019905e-12 0.4521651 0.616 0.334 1.604143e-07 1 ## Ramp2 4.146672e-12 1.3163955 0.204 0.067 1.654730e-07 1 ## Sh3yl11 4.183451e-12 0.4472919 0.584 0.326 1.669406e-07 1 ## Epsti1 4.192336e-12 0.5525731 0.498 0.248 1.672952e-07 1 ## Smim291 4.222122e-12 0.5423498 0.620 0.338 1.684838e-07 1 ## Timm17a1 4.229019e-12 0.4936611 0.755 0.457 1.687590e-07 1 ## Smpdl3a 4.247333e-12 0.2679753 0.902 0.577 1.694898e-07 1 ## Marchf91 4.262578e-12 0.6761391 0.473 0.231 1.700982e-07 1 ## Prrc1 4.300195e-12 0.3899043 0.771 0.464 1.715993e-07 1 ## Ube2h 4.309056e-12 0.4856221 0.784 0.491 1.719529e-07 1 ## Mocs1 4.453324e-12 0.1568659 0.461 0.221 1.777099e-07 1 ## Chchd6 4.457519e-12 1.0157227 0.290 0.117 1.778773e-07 1 ## Ndufb101 4.524308e-12 0.3417143 0.963 0.705 1.805425e-07 1 ## Frmd6 4.595446e-12 1.9561858 0.102 0.019 1.833813e-07 1 ## Gm41753 4.627266e-12 2.3118866 0.098 0.017 1.846511e-07 1 ## Pcmt1 4.703944e-12 0.4603870 0.759 0.479 1.877109e-07 1 ## Hras1 4.833440e-12 0.4563556 0.853 0.586 1.928784e-07 1 ## Desi2 4.861569e-12 0.5384694 0.641 0.350 1.940009e-07 1 ## Fes 5.037822e-12 1.1320071 0.216 0.071 2.010343e-07 1 ## F8a 5.098323e-12 1.0498064 0.343 0.155 2.034486e-07 1 ## Zfp81 5.213197e-12 1.0872345 0.273 0.108 2.080326e-07 1 ## Fndc3a 5.240850e-12 0.3238510 0.980 0.757 2.091361e-07 1 ## Unc119 5.285122e-12 0.9055025 0.302 0.122 2.109028e-07 1 ## Avpi1 5.544833e-12 1.2972538 0.253 0.098 2.212666e-07 1 ## Polr2g1 5.569713e-12 0.4991528 0.824 0.542 2.222594e-07 1 ## Tle6 5.645655e-12 0.5734705 0.539 0.278 2.252899e-07 1 ## Mvk 5.815480e-12 0.5927440 0.371 0.169 2.320667e-07 1 ## Klhdc1 5.863735e-12 1.2053372 0.208 0.070 2.339924e-07 1 ## Extl2 5.950176e-12 0.7136700 0.265 0.099 2.374418e-07 1 ## Sort11 6.087303e-12 0.4371954 0.812 0.526 2.429138e-07 1 ## Ing1 6.362355e-12 0.4779014 0.690 0.423 2.538898e-07 1 ## Gnptg1 6.461344e-12 0.4685212 0.906 0.646 2.578399e-07 1 ## Tmem2051 6.630504e-12 0.3359028 0.894 0.585 2.645903e-07 1 ## Gm45998 6.733962e-12 0.8979996 0.294 0.125 2.687188e-07 1 ## Hacd1 6.884692e-12 0.1624880 0.686 0.394 2.747336e-07 1 ## Tma71 6.987320e-12 0.3370346 0.980 0.767 2.788290e-07 1 ## Arhgef9 7.078376e-12 1.3320556 0.163 0.045 2.824626e-07 1 ## Laptm4a1 7.189831e-12 0.3594967 0.992 0.760 2.869102e-07 1 ## Anapc11 7.361557e-12 0.4419306 0.637 0.363 2.937629e-07 1 ## Arhgef10l 7.374719e-12 1.4832301 0.143 0.037 2.942882e-07 1 ## Nhlh2 7.416812e-12 4.6747151 0.033 0.000 2.959679e-07 1 ## Mobp 7.416812e-12 5.0726781 0.033 0.000 2.959679e-07 1 ## Cmya5 7.416812e-12 4.5796609 0.033 0.000 2.959679e-07 1 ## Vbp11 7.617592e-12 0.4583413 0.747 0.476 3.039800e-07 1 ## P3h1 7.624334e-12 2.5170774 0.078 0.010 3.042490e-07 1 ## Dpyd 7.696497e-12 0.3681952 0.849 0.563 3.071287e-07 1 ## Mmp24 8.073119e-12 1.0559458 0.180 0.053 3.221578e-07 1 ## Atl2 8.171472e-12 0.4115019 0.771 0.468 3.260826e-07 1 ## Cmbl 8.209194e-12 0.4002709 0.449 0.217 3.275879e-07 1 ## Zfp941 8.354007e-12 1.8170131 0.114 0.024 3.333667e-07 1 ## Adk 8.466754e-12 0.5071475 0.478 0.238 3.378658e-07 1 ## Bola3 8.486358e-12 0.2627641 0.731 0.426 3.386481e-07 1 ## Gbp9 8.534015e-12 0.9375579 0.269 0.103 3.405499e-07 1 ## Ctps2 8.577821e-12 0.7841019 0.445 0.225 3.422980e-07 1 ## Tm7sf3 8.651639e-12 0.6678569 0.429 0.210 3.452436e-07 1 ## Zswim5 8.765818e-12 1.3073705 0.233 0.087 3.498000e-07 1 ## Igdcc4 8.821330e-12 3.2625084 0.065 0.007 3.520152e-07 1 ## Fam229b 8.949382e-12 1.9206823 0.127 0.029 3.571251e-07 1 ## Prkaa11 9.097373e-12 0.3440481 0.755 0.454 3.630307e-07 1 ## Sdhaf1 9.296044e-12 0.7364189 0.469 0.238 3.709586e-07 1 ## 1300002E11Rik 9.314180e-12 0.4091266 0.759 0.457 3.716823e-07 1 ## 2810001A02Rik 9.568052e-12 0.6590943 0.612 0.347 3.818131e-07 1 ## Cox7c 9.688974e-12 0.1770063 1.000 0.891 3.866385e-07 1 ## Setd31 9.741857e-12 0.4136564 0.878 0.603 3.887488e-07 1 ## Ivd 9.840591e-12 0.5685786 0.494 0.257 3.926888e-07 1 ## Tcp11 1.031243e-11 0.2972327 0.918 0.615 4.115176e-07 1 ## Ndrg4 1.032921e-11 2.7254064 0.065 0.007 4.121872e-07 1 ## Ephb2 1.064251e-11 0.6347943 0.388 0.182 4.246894e-07 1 ## Gm14279 1.071993e-11 0.3725916 0.808 0.521 4.277789e-07 1 ## Dpy19l1 1.089453e-11 0.7257885 0.192 0.062 4.347464e-07 1 ## Lefty1 1.116045e-11 1.1405788 0.257 0.098 4.453579e-07 1 ## Dad11 1.133549e-11 0.3542323 0.996 0.800 4.523429e-07 1 ## Dbndd2 1.136400e-11 0.6862558 0.506 0.268 4.534806e-07 1 ## Hfe 1.138372e-11 0.8716553 0.192 0.062 4.542675e-07 1 ## Zfp932 1.148850e-11 0.7688337 0.367 0.168 4.584487e-07 1 ## Sil1 1.150334e-11 0.6397544 0.543 0.296 4.590408e-07 1 ## Mrpl201 1.168692e-11 0.3420007 0.914 0.604 4.663666e-07 1 ## Hcar11 1.196561e-11 0.9096359 0.253 0.094 4.774876e-07 1 ## Usp8 1.284738e-11 0.4560633 0.796 0.505 5.126747e-07 1 ## Slc66a3 1.288079e-11 1.2066890 0.253 0.096 5.140080e-07 1 ## Drg11 1.290948e-11 0.4458833 0.624 0.349 5.151526e-07 1 ## Rapgef4 1.325281e-11 0.2900203 0.669 0.385 5.288533e-07 1 ## Zfp362 1.344910e-11 0.6173656 0.359 0.157 5.366862e-07 1 ## Prelid1 1.353390e-11 0.2112900 0.980 0.737 5.400701e-07 1 ## Gm54305 1.377729e-11 3.8539253 0.037 0.001 5.497828e-07 1 ## Gadd45g1 1.382235e-11 0.4873222 0.755 0.482 5.515809e-07 1 ## Serp21 1.416491e-11 0.9533157 0.212 0.071 5.652508e-07 1 ## Acyp1 1.427146e-11 0.7202903 0.547 0.305 5.695028e-07 1 ## 2200002J24Rik 1.481071e-11 2.6666743 0.090 0.015 5.910213e-07 1 ## Rxylt1 1.487215e-11 0.6710086 0.490 0.254 5.934730e-07 1 ## Bag4 1.499780e-11 0.4275795 0.682 0.382 5.984871e-07 1 ## Ncstn1 1.507973e-11 0.5296502 0.788 0.526 6.017568e-07 1 ## Mtfr1l 1.515734e-11 0.5123844 0.535 0.289 6.048537e-07 1 ## Gm16124 1.617584e-11 1.2408788 0.241 0.089 6.454969e-07 1 ## Tmem237 1.623220e-11 0.9869367 0.384 0.185 6.477458e-07 1 ## Dlat 1.637768e-11 0.6247726 0.396 0.185 6.535513e-07 1 ## Mettl9 1.657247e-11 0.4504948 0.731 0.444 6.613245e-07 1 ## 2310009B15Rik 1.658900e-11 0.9115949 0.371 0.176 6.619841e-07 1 ## Rhou 1.660637e-11 0.2289202 0.914 0.605 6.626772e-07 1 ## Mtif2 1.718848e-11 0.4176885 0.706 0.410 6.859062e-07 1 ## Hmg20a 1.774191e-11 0.6677504 0.376 0.176 7.079910e-07 1 ## Btf3l4 1.789740e-11 0.4442260 0.767 0.475 7.141956e-07 1 ## Hsd17b10 1.791865e-11 0.3119018 0.612 0.321 7.150438e-07 1 ## Edaradd 1.793529e-11 2.1976100 0.073 0.010 7.157078e-07 1 ## Ampd3 1.793614e-11 0.7110260 0.351 0.161 7.157415e-07 1 ## Gmfb1 1.828692e-11 0.3794661 0.706 0.419 7.297397e-07 1 ## St18 1.844751e-11 0.1282423 0.763 0.430 7.361480e-07 1 ## Armcx2 1.859142e-11 2.7566040 0.086 0.014 7.418907e-07 1 ## Ubxn1 1.860624e-11 0.2987146 0.971 0.714 7.424822e-07 1 ## Kcna4 1.964263e-11 1.7505610 0.073 0.010 7.838392e-07 1 ## Ecd 2.035046e-11 0.6494672 0.486 0.259 8.120850e-07 1 ## Gpr108 2.037104e-11 0.3834686 0.616 0.330 8.129064e-07 1 ## Socs1 2.039943e-11 1.8523777 0.114 0.025 8.140391e-07 1 ## C1ra 2.082792e-11 3.3742740 0.061 0.006 8.311380e-07 1 ## Ankra21 2.088807e-11 0.3427112 0.800 0.509 8.335384e-07 1 ## Fkbp3 2.118308e-11 0.3568861 0.739 0.416 8.453109e-07 1 ## Aldh18a1 2.156189e-11 0.3935204 0.616 0.340 8.604274e-07 1 ## Gm38952 2.159860e-11 0.7846261 0.196 0.063 8.618923e-07 1 ## Klc3 2.172040e-11 0.4660758 0.473 0.226 8.667524e-07 1 ## Sra11 2.176953e-11 0.4618229 0.886 0.590 8.687131e-07 1 ## Thumpd1 2.306440e-11 0.3099376 0.494 0.253 9.203850e-07 1 ## Adgrl2 2.315518e-11 0.8296751 0.331 0.151 9.240075e-07 1 ## Gm19689 2.316382e-11 1.8381372 0.073 0.010 9.243522e-07 1 ## Slc23a2 2.340697e-11 0.7233541 0.286 0.118 9.340553e-07 1 ## Tmem35b 2.407331e-11 0.9965758 0.310 0.134 9.606456e-07 1 ## Srpk2 2.495861e-11 0.3057029 0.792 0.498 9.959731e-07 1 ## Txndc16 2.641614e-11 1.2530881 0.180 0.057 1.054136e-06 1 ## Tuba1a1 2.657764e-11 0.2635285 0.914 0.631 1.060581e-06 1 ## Gpr180 2.667584e-11 0.8576956 0.376 0.178 1.064499e-06 1 ## Ctxn1 2.673827e-11 1.0108584 0.261 0.102 1.066991e-06 1 ## Pcmtd11 2.694776e-11 0.3682614 0.861 0.588 1.075350e-06 1 ## Rtraf 2.718152e-11 0.2705425 0.890 0.580 1.084679e-06 1 ## Gm54268 2.750663e-11 0.5312748 0.367 0.171 1.097652e-06 1 ## Zbtb7c 2.773957e-11 1.1510455 0.106 0.022 1.106948e-06 1 ## Fgl1 2.815606e-11 2.6471000 0.106 0.022 1.123568e-06 1 ## Cyp2c651 2.832761e-11 0.3076328 0.898 0.604 1.130413e-06 1 ## Gm51806 2.837399e-11 0.2529735 0.486 0.245 1.132264e-06 1 ## Timm10b1 2.878381e-11 0.4601028 0.873 0.567 1.148618e-06 1 ## Atp5pb 2.995181e-11 0.1010764 0.996 0.824 1.195227e-06 1 ## Gm44008 3.029776e-11 1.0045521 0.192 0.062 1.209032e-06 1 ## Msto1 3.063337e-11 1.0426509 0.257 0.102 1.222425e-06 1 ## Mapk31 3.122717e-11 0.2880787 1.000 0.854 1.246120e-06 1 ## Coro2b 3.135183e-11 1.8651334 0.102 0.020 1.251095e-06 1 ## Haghl 3.201869e-11 0.5847096 0.449 0.226 1.277706e-06 1 ## Prkar1a1 3.248898e-11 0.3263771 0.992 0.799 1.296473e-06 1 ## Prr15 3.306988e-11 0.3289399 0.457 0.225 1.319654e-06 1 ## 1810008I18Rik 3.364310e-11 1.5374612 0.127 0.031 1.342528e-06 1 ## Kdelr21 3.397403e-11 0.3545932 0.980 0.772 1.355734e-06 1 ## Sec22b 3.399860e-11 0.4481807 0.633 0.375 1.356714e-06 1 ## Pts 3.474863e-11 0.3978814 0.796 0.495 1.386644e-06 1 ## Trappc6a1 3.487291e-11 0.3666830 0.841 0.523 1.391603e-06 1 ## Hsdl2 3.490777e-11 0.6708478 0.424 0.210 1.392994e-06 1 ## E2f2 3.491103e-11 0.6455032 0.482 0.252 1.393125e-06 1 ## Emc101 3.507295e-11 0.3878863 0.922 0.681 1.399586e-06 1 ## Gng2 3.538632e-11 0.9308613 0.257 0.100 1.412091e-06 1 ## Timm231 3.583375e-11 0.3560900 0.776 0.475 1.429946e-06 1 ## Spcs3 3.653395e-11 0.3179113 0.820 0.514 1.457887e-06 1 ## A1cf 3.684835e-11 0.2972336 0.939 0.700 1.470433e-06 1 ## Mrps17 3.742153e-11 0.3903226 0.682 0.375 1.493306e-06 1 ## Crlf2 3.746530e-11 0.9876901 0.367 0.173 1.495053e-06 1 ## Ptpmt1 3.808956e-11 0.3282161 0.673 0.373 1.519964e-06 1 ## Med30 3.844022e-11 0.5280627 0.535 0.288 1.533957e-06 1 ## Abhd16a1 3.908277e-11 0.3806088 0.796 0.500 1.559598e-06 1 ## Erh 4.281120e-11 0.2109220 0.906 0.587 1.708381e-06 1 ## Tspyl4 4.340877e-11 0.4927954 0.424 0.204 1.732227e-06 1 ## Lyrm9 4.351328e-11 0.4777314 0.612 0.352 1.736397e-06 1 ## Rab11a1 4.488557e-11 0.3142423 0.890 0.644 1.791159e-06 1 ## Prdx4 4.502340e-11 0.4435258 0.335 0.144 1.796659e-06 1 ## Nxn 4.597488e-11 1.6899727 0.131 0.033 1.834627e-06 1 ## Zfp74 4.796812e-11 0.6897567 0.306 0.126 1.914168e-06 1 ## Tmem126a1 5.067842e-11 0.4699246 0.751 0.474 2.022322e-06 1 ## Def8 5.150446e-11 0.7580517 0.367 0.171 2.055286e-06 1 ## Ngrn 5.217370e-11 0.5930502 0.584 0.325 2.081991e-06 1 ## Tyw5 5.266106e-11 1.1287938 0.253 0.102 2.101440e-06 1 ## Rapgef5 5.307225e-11 0.5624185 0.347 0.157 2.117848e-06 1 ## Gm12343 5.337760e-11 2.1555302 0.086 0.015 2.130033e-06 1 ## 1600027J07Rik 5.402866e-11 2.9366413 0.057 0.006 2.156014e-06 1 ## Atp6v0e21 5.477105e-11 0.5644987 0.706 0.428 2.185639e-06 1 ## Nr1i2 5.482064e-11 0.4904788 0.567 0.307 2.187618e-06 1 ## Bri3 5.578940e-11 0.3126667 0.935 0.621 2.226276e-06 1 ## Smim19 5.706586e-11 0.6482361 0.473 0.254 2.277213e-06 1 ## Slc9a7 5.743924e-11 0.8919611 0.314 0.136 2.292113e-06 1 ## Bcor 5.807951e-11 0.3979342 0.539 0.285 2.317663e-06 1 ## Cdkn1a 5.971825e-11 0.2607791 0.996 0.850 2.383057e-06 1 ## H2-T231 6.016203e-11 0.3130594 0.980 0.741 2.400766e-06 1 ## Gas7 6.099768e-11 3.0689580 0.057 0.006 2.434112e-06 1 ## Ppme1 6.139360e-11 0.3531882 0.722 0.448 2.449912e-06 1 ## Qsox2 6.180251e-11 0.4677867 0.449 0.231 2.466229e-06 1 ## Mff 6.338402e-11 0.4336457 0.763 0.488 2.529339e-06 1 ## Alkbh6 6.442116e-11 0.6331985 0.584 0.324 2.570726e-06 1 ## Gm33636 6.464791e-11 3.1690202 0.057 0.006 2.579775e-06 1 ## Gm343371 6.503387e-11 0.9835907 0.241 0.087 2.595176e-06 1 ## Bmyc 6.526125e-11 0.5738240 0.551 0.301 2.604250e-06 1 ## Lancl1 6.538221e-11 0.8907062 0.371 0.179 2.609077e-06 1 ## Triobp1 6.582190e-11 0.4760614 0.645 0.379 2.626623e-06 1 ## Ssna11 6.595326e-11 0.5480108 0.714 0.418 2.631865e-06 1 ## Gm465601 6.636422e-11 0.9448391 0.257 0.100 2.648264e-06 1 ## Scnm1 6.906950e-11 0.7489678 0.396 0.196 2.756219e-06 1 ## Tmem258 7.181668e-11 0.3137027 0.939 0.667 2.865845e-06 1 ## Nr2f2 7.289134e-11 0.4501957 0.461 0.237 2.908729e-06 1 ## Ube2d2a1 7.489236e-11 0.3477639 0.902 0.639 2.988580e-06 1 ## Tsnax 7.699972e-11 0.4714348 0.616 0.352 3.072674e-06 1 ## 2310022B05Rik 7.878430e-11 0.6134431 0.514 0.289 3.143887e-06 1 ## Pigyl 7.997772e-11 0.6084569 0.588 0.326 3.191511e-06 1 ## Lsm3 8.054937e-11 0.3912923 0.576 0.314 3.214323e-06 1 ## Frat2 8.339680e-11 0.6689472 0.518 0.280 3.327949e-06 1 ## Cgn 8.398676e-11 0.1180243 0.661 0.368 3.351492e-06 1 ## Zfp704 8.416566e-11 0.2350733 0.661 0.368 3.358631e-06 1 ## Myl12b1 8.523960e-11 0.3010303 0.984 0.800 3.401486e-06 1 ## Zfp8711 8.531912e-11 0.4118633 0.914 0.654 3.404660e-06 1 ## Cd200 8.576638e-11 0.2869402 0.694 0.442 3.422508e-06 1 ## Fem1b1 8.599840e-11 0.4334490 0.714 0.430 3.431766e-06 1 ## Uqcc51 8.757087e-11 0.4048604 0.731 0.439 3.494515e-06 1 ## Tm2d11 8.774610e-11 0.4082724 0.833 0.543 3.501508e-06 1 ## Ntsr2 8.846060e-11 1.3485502 0.147 0.041 3.530020e-06 1 ## 1500011B03Rik1 9.148599e-11 0.4697932 0.596 0.338 3.650748e-06 1 ## Cox14 9.255655e-11 0.3016949 0.922 0.665 3.693469e-06 1 ## Eif4a2 9.348016e-11 0.3623994 0.963 0.758 3.730326e-06 1 ## Lipc1 9.559739e-11 0.9579202 0.159 0.046 3.814814e-06 1 ## Slc44a11 9.601026e-11 0.3538777 0.837 0.544 3.831289e-06 1 ## Rprd1a 9.707851e-11 0.6107305 0.531 0.291 3.873918e-06 1 ## Myc 9.709907e-11 0.9416573 0.220 0.082 3.874739e-06 1 ## Fam83b 9.878969e-11 0.2295094 0.571 0.326 3.942203e-06 1 ## Tmed4 9.971064e-11 0.3419768 0.882 0.617 3.978953e-06 1 ## Atp2a21 1.007410e-10 0.2520579 0.980 0.826 4.020069e-06 1 ## Cul7 1.020540e-10 0.6781794 0.335 0.147 4.072466e-06 1 ## Pygb 1.025622e-10 0.7550366 0.204 0.074 4.092745e-06 1 ## Ndufab1 1.103632e-10 0.1460378 0.939 0.635 4.404045e-06 1 ## Gemin5 1.106525e-10 1.0191362 0.224 0.087 4.415588e-06 1 ## Tmem2381 1.108084e-10 0.3409983 0.914 0.660 4.421811e-06 1 ## Mtmr9 1.141039e-10 0.5310410 0.551 0.308 4.553316e-06 1 ## Zdhhc17 1.179225e-10 0.8893277 0.367 0.178 4.705698e-06 1 ## Sigmar1 1.180331e-10 0.7886393 0.396 0.198 4.710109e-06 1 ## Gtf2e2 1.189686e-10 0.3649296 0.735 0.445 4.747442e-06 1 ## Dbp 1.223858e-10 1.0586680 0.241 0.094 4.883807e-06 1 ## Prmt11 1.228493e-10 0.3745206 0.739 0.436 4.902303e-06 1 ## Tmem14a 1.310309e-10 1.2792517 0.229 0.087 5.228789e-06 1 ## Ldaf1 1.311227e-10 0.8167213 0.396 0.198 5.232453e-06 1 ## Lekr1 1.312384e-10 1.1205353 0.184 0.061 5.237068e-06 1 ## Zfp318 1.329769e-10 0.4341549 0.547 0.305 5.306444e-06 1 ## Eid2 1.342959e-10 0.5842043 0.420 0.207 5.359079e-06 1 ## Gal 1.374368e-10 3.0210636 0.065 0.008 5.484416e-06 1 ## Vamp21 1.375302e-10 0.4099324 0.808 0.526 5.488144e-06 1 ## Gm40743 1.398417e-10 1.0544771 0.102 0.022 5.580383e-06 1 ## 2310010J17Rik 1.436465e-10 1.1562360 0.229 0.088 5.732214e-06 1 ## Gtf2f1 1.470815e-10 0.3352968 0.669 0.391 5.869288e-06 1 ## Creb3 1.475106e-10 0.5728804 0.612 0.358 5.886410e-06 1 ## Cmtm8 1.488198e-10 2.7389997 0.069 0.010 5.938653e-06 1 ## Cyp3a16 1.503505e-10 4.3353537 0.029 0.000 5.999737e-06 1 ## Gm53937 1.503505e-10 4.3083990 0.029 0.000 5.999737e-06 1 ## Vps291 1.564414e-10 0.3726010 0.833 0.557 6.242795e-06 1 ## Ino80d 1.582264e-10 0.3780226 0.816 0.544 6.314023e-06 1 ## Gal3st1 1.582737e-10 0.4520799 0.498 0.268 6.315912e-06 1 ## Spryd7 1.605511e-10 0.5345540 0.478 0.254 6.406791e-06 1 ## Atp6ap11 1.606602e-10 0.3675173 0.898 0.627 6.411143e-06 1 ## Micu31 1.627940e-10 0.7821845 0.412 0.212 6.496294e-06 1 ## Hint31 1.650571e-10 0.3914784 0.747 0.471 6.586602e-06 1 ## Cep112 1.678528e-10 1.7882890 0.106 0.024 6.698165e-06 1 ## Polb 1.700649e-10 0.5933454 0.527 0.287 6.786440e-06 1 ## Rbm43 1.711865e-10 1.4199901 0.188 0.066 6.831196e-06 1 ## Syt14 1.739999e-10 0.4992221 0.453 0.229 6.943467e-06 1 ## Gnb21 1.778156e-10 0.3024739 0.992 0.846 7.095732e-06 1 ## Eef1akmt2 1.795826e-10 0.6111655 0.400 0.201 7.166245e-06 1 ## Meiob 1.813927e-10 2.7275393 0.041 0.002 7.238475e-06 1 ## Cnst 1.850659e-10 0.3371284 0.498 0.250 7.385055e-06 1 ## Tmem276 1.906614e-10 0.5296628 0.404 0.199 7.608343e-06 1 ## Nudt10 1.915375e-10 0.8966237 0.273 0.116 7.643305e-06 1 ## Polr2c 1.931692e-10 0.4671765 0.690 0.435 7.708416e-06 1 ## Trappc2b 1.949481e-10 0.3425968 0.698 0.420 7.779402e-06 1 ## Jtb 1.957355e-10 0.3770129 0.796 0.526 7.810824e-06 1 ## Sar1a1 1.981327e-10 0.3502313 0.837 0.564 7.906484e-06 1 ## Suclg1 1.998239e-10 0.1850867 0.951 0.665 7.973974e-06 1 ## A930038B10Rik1 2.042249e-10 1.2556744 0.155 0.046 8.149594e-06 1 ## Myl12a 2.047116e-10 0.3079692 0.767 0.468 8.169016e-06 1 ## Itfg11 2.078099e-10 0.3638311 0.808 0.552 8.292655e-06 1 ## Nbas1 2.101637e-10 0.3209280 0.796 0.525 8.386584e-06 1 ## Man2a2 2.144526e-10 0.4768077 0.416 0.201 8.557730e-06 1 ## Gpr85 2.185673e-10 1.4854032 0.139 0.039 8.721928e-06 1 ## Rex1bd1 2.278858e-10 0.4162021 0.841 0.552 9.093781e-06 1 ## Rin31 2.343254e-10 0.5407090 0.343 0.150 9.350754e-06 1 ## Krt1 2.370471e-10 4.3213509 0.037 0.001 9.459363e-06 1 ## Ninj1 2.407932e-10 0.4483986 0.637 0.375 9.608854e-06 1 ## Dll41 2.432178e-10 0.3860422 0.502 0.269 9.705607e-06 1 ## Gm47095 2.434111e-10 4.0866414 0.037 0.001 9.713319e-06 1 ## Tenm2 2.499419e-10 2.9916761 0.037 0.001 9.973930e-06 1 ## R3hdm2 2.513830e-10 0.3043125 0.882 0.600 1.003144e-05 1 ## Fdx1 2.543462e-10 0.3619782 0.502 0.268 1.014969e-05 1 ## Etv4 2.570590e-10 1.1482523 0.180 0.060 1.025794e-05 1 ## Thap3 2.624301e-10 0.5537422 0.510 0.272 1.047227e-05 1 ## Rufy2 2.672643e-10 0.3840619 0.486 0.252 1.066518e-05 1 ## Ldlrad4 2.685805e-10 0.2920433 0.559 0.307 1.071770e-05 1 ## Sumf2 2.818785e-10 1.1584293 0.196 0.069 1.124836e-05 1 ## Cadps1 2.870150e-10 0.2773093 0.939 0.668 1.145333e-05 1 ## Lyl1 2.874935e-10 2.0839782 0.135 0.038 1.147243e-05 1 ## Gm13594 2.916610e-10 1.7136023 0.114 0.028 1.163873e-05 1 ## Septin10 2.982011e-10 0.6802012 0.347 0.164 1.189971e-05 1 ## Ppp2ca1 2.997680e-10 0.3338112 0.939 0.660 1.196224e-05 1 ## Smim8 3.018686e-10 0.5205684 0.559 0.323 1.204607e-05 1 ## Nkiras2 3.073794e-10 0.3891120 0.608 0.368 1.226597e-05 1 ## Cby1 3.078198e-10 0.8190935 0.327 0.152 1.228355e-05 1 ## Snrnp271 3.109401e-10 0.4544280 0.743 0.461 1.240807e-05 1 ## Agpat5 3.171976e-10 0.5225488 0.563 0.326 1.265777e-05 1 ## Ebna1bp2 3.174147e-10 0.3894632 0.682 0.388 1.266643e-05 1 ## Aph1b 3.182201e-10 1.0795129 0.229 0.088 1.269857e-05 1 ## H1f10 3.213904e-10 0.5999772 0.637 0.391 1.282508e-05 1 ## Sgms1os1 3.240470e-10 0.8856677 0.302 0.134 1.293109e-05 1 ## Ndufb3 3.248447e-10 0.2279512 0.955 0.681 1.296293e-05 1 ## Gabarapl1 3.257671e-10 0.4179620 0.551 0.307 1.299974e-05 1 ## Idi11 3.260932e-10 0.3014075 0.657 0.385 1.301275e-05 1 ## Card191 3.265094e-10 0.3564105 0.808 0.531 1.302936e-05 1 ## Atp5mg1 3.296404e-10 0.2452655 1.000 0.840 1.315430e-05 1 ## Cd22 3.303396e-10 1.5916312 0.110 0.026 1.318220e-05 1 ## Bola21 3.311270e-10 0.4456007 0.755 0.490 1.321362e-05 1 ## Sar1b1 3.331751e-10 0.3128232 0.898 0.614 1.329535e-05 1 ## Eef2kmt 3.446598e-10 0.4605658 0.494 0.269 1.375365e-05 1 ## Magoh 3.468942e-10 0.4208756 0.718 0.428 1.384281e-05 1 ## Siah21 3.558579e-10 0.3660096 0.580 0.328 1.420051e-05 1 ## Galnt31 3.580927e-10 0.3961862 0.665 0.411 1.428969e-05 1 ## Aldh6a1 3.641244e-10 0.7237804 0.347 0.166 1.453038e-05 1 ## Sacs 3.676568e-10 1.7071690 0.090 0.017 1.467135e-05 1 ## Ss18l2 3.847034e-10 0.5205972 0.596 0.352 1.535159e-05 1 ## Bloc1s2 3.855880e-10 0.3986512 0.616 0.342 1.538689e-05 1 ## Psph 3.869777e-10 1.0829937 0.269 0.117 1.544234e-05 1 ## Txndc121 3.876789e-10 0.6750584 0.547 0.312 1.547032e-05 1 ## Atp9a1 3.904364e-10 0.3723116 0.894 0.666 1.558036e-05 1 ## Mycbp2 4.259924e-10 0.2660344 0.857 0.603 1.699923e-05 1 ## Actr101 4.282236e-10 0.5109977 0.796 0.558 1.708826e-05 1 ## Coq2 4.335555e-10 0.4304434 0.641 0.392 1.730103e-05 1 ## Macrod2 4.600371e-10 1.2762927 0.171 0.057 1.835778e-05 1 ## Adcy2 4.693321e-10 0.1996281 0.673 0.380 1.872870e-05 1 ## Wdr6 4.740821e-10 0.5017248 0.616 0.366 1.891825e-05 1 ## Pcyox1l1 4.759443e-10 0.5661057 0.449 0.233 1.899256e-05 1 ## Fam8a1 4.795204e-10 0.3483952 0.649 0.399 1.913526e-05 1 ## Arl21 4.816757e-10 0.5140030 0.690 0.444 1.922127e-05 1 ## Gm40922 4.879419e-10 2.3677349 0.073 0.012 1.947132e-05 1 ## Wdr83 4.907451e-10 0.6324188 0.416 0.217 1.958318e-05 1 ## 2410006H16Rik 4.955131e-10 0.1007200 0.755 0.449 1.977345e-05 1 ## Ttc331 4.960022e-10 0.4956320 0.620 0.379 1.979297e-05 1 ## H2az2 5.081945e-10 0.1029780 0.914 0.616 2.027950e-05 1 ## Vgll4 5.091470e-10 0.1467012 0.449 0.222 2.031751e-05 1 ## Gm34147 5.100859e-10 2.3226825 0.069 0.010 2.035498e-05 1 ## Tyw1 5.126579e-10 0.5567164 0.351 0.169 2.045762e-05 1 ## Pcsk11 5.299486e-10 0.3117848 0.996 0.846 2.114760e-05 1 ## Spg71 5.352777e-10 0.4002144 0.759 0.512 2.136026e-05 1 ## Gm20208 5.407058e-10 2.2422747 0.065 0.009 2.157686e-05 1 ## F11r1 5.425445e-10 0.3503939 0.735 0.457 2.165024e-05 1 ## Tmem161a 5.566879e-10 0.4960223 0.502 0.277 2.221463e-05 1 ## Zmat21 5.621300e-10 0.3528489 0.861 0.572 2.243180e-05 1 ## 9030622O22Rik1 5.656072e-10 1.0532330 0.176 0.059 2.257055e-05 1 ## Cutal1 5.721020e-10 0.5466858 0.392 0.188 2.282973e-05 1 ## Zhx11 5.724807e-10 0.4665524 0.588 0.347 2.284484e-05 1 ## Ralgps1 5.811457e-10 0.5973575 0.229 0.089 2.319062e-05 1 ## Auh 5.833884e-10 0.4538808 0.665 0.398 2.328011e-05 1 ## F21 5.869386e-10 0.3053268 0.588 0.312 2.342179e-05 1 ## Akr1c121 5.878984e-10 0.2878652 0.902 0.622 2.346009e-05 1 ## Timmdc1 5.964144e-10 0.3519553 0.457 0.246 2.379992e-05 1 ## Utp3 6.005787e-10 0.4314419 0.629 0.361 2.396609e-05 1 ## Mrps24 6.008765e-10 0.2466080 0.898 0.609 2.397798e-05 1 ## 2310015A10Rik 6.024794e-10 1.1742426 0.180 0.061 2.404194e-05 1 ## D730003I15Rik 6.194225e-10 0.6694356 0.302 0.136 2.471805e-05 1 ## Synrg1 6.251134e-10 0.3311110 0.845 0.586 2.494515e-05 1 ## Cdk2ap11 6.411606e-10 0.3208092 0.865 0.561 2.558551e-05 1 ## Il11ra1 6.421485e-10 0.8101261 0.380 0.192 2.562493e-05 1 ## Foxo4 6.692434e-10 0.7893129 0.355 0.177 2.670616e-05 1 ## Psmb51 6.769486e-10 0.3313752 0.935 0.687 2.701364e-05 1 ## Unkl 6.827373e-10 0.8274352 0.192 0.068 2.724463e-05 1 ## Pja21 6.867218e-10 0.3614045 0.841 0.583 2.740364e-05 1 ## Luc7l2 6.946831e-10 0.1910211 0.992 0.862 2.772133e-05 1 ## Gm41516 7.020805e-10 2.6209527 0.053 0.006 2.801652e-05 1 ## Dnajc21 7.052829e-10 0.4263568 0.690 0.399 2.814431e-05 1 ## Gm30173 7.088922e-10 3.4647057 0.045 0.003 2.828834e-05 1 ## Gpd1l 7.394017e-10 0.5511925 0.302 0.136 2.950582e-05 1 ## Ypel5 7.504468e-10 0.3579607 0.539 0.294 2.994658e-05 1 ## Smpd31 7.631720e-10 0.3283817 0.947 0.739 3.045438e-05 1 ## 1700110K17Rik 7.663621e-10 1.6160242 0.143 0.043 3.058168e-05 1 ## Gorasp21 7.851665e-10 0.2274899 0.767 0.479 3.133207e-05 1 ## Plpp5 7.889990e-10 0.4054503 0.551 0.315 3.148500e-05 1 ## Ammecr1 7.890702e-10 0.7654546 0.376 0.194 3.148784e-05 1 ## Dnajb141 7.908966e-10 0.4572716 0.620 0.374 3.156073e-05 1 ## Tmem30a1 8.042878e-10 0.3808076 0.935 0.725 3.209510e-05 1 ## Rab13 8.137287e-10 0.8091421 0.322 0.152 3.247185e-05 1 ## Fam3c1 8.205804e-10 0.2956409 0.657 0.400 3.274526e-05 1 ## Hsbp1l1 8.353302e-10 0.7300601 0.335 0.157 3.333385e-05 1 ## Chmp1b2 8.427649e-10 0.5546826 0.518 0.294 3.363053e-05 1 ## Galnt14 8.477484e-10 0.2622494 0.894 0.596 3.382940e-05 1 ## Gm51438 8.571332e-10 0.6940750 0.237 0.094 3.420390e-05 1 ## Vma211 8.610605e-10 0.4026618 0.608 0.356 3.436062e-05 1 ## Eri3 8.671851e-10 0.3467944 0.612 0.365 3.460502e-05 1 ## Gstm71 8.859821e-10 0.6560419 0.416 0.215 3.535511e-05 1 ## Cebpg1 8.907972e-10 0.3838903 0.820 0.578 3.554726e-05 1 ## Dtna1 8.985601e-10 1.5639873 0.106 0.025 3.585704e-05 1 ## Dnase2a 9.024734e-10 1.3485713 0.114 0.029 3.601320e-05 1 ## Oas1a 9.032547e-10 1.0882326 0.176 0.061 3.604438e-05 1 ## Usp291 9.299346e-10 0.3792069 0.437 0.222 3.710904e-05 1 ## Adal 9.327409e-10 0.8348268 0.253 0.107 3.722103e-05 1 ## Neu2 9.353898e-10 3.2717755 0.057 0.007 3.732673e-05 1 ## Glg11 9.439237e-10 0.3226506 0.951 0.732 3.766727e-05 1 ## Pdia6 9.478215e-10 0.3307273 0.914 0.655 3.782282e-05 1 ## Btbd11 9.581574e-10 0.3958508 0.771 0.497 3.823527e-05 1 ## Cdc23 9.588924e-10 0.6490970 0.396 0.203 3.826460e-05 1 ## Rsu1 9.606152e-10 1.1075657 0.253 0.110 3.833335e-05 1 ## Cib2 9.704421e-10 0.8899682 0.220 0.085 3.872549e-05 1 ## Pold2 9.734940e-10 0.4603906 0.453 0.238 3.884728e-05 1 ## Slc66a21 9.838135e-10 0.4823443 0.433 0.222 3.925908e-05 1 ## Ube2l6 9.993866e-10 0.7367800 0.351 0.171 3.988052e-05 1 ## Pank31 1.004627e-09 0.3589641 0.841 0.578 4.008964e-05 1 ## Mrpl12 1.015097e-09 0.4268553 0.690 0.414 4.050746e-05 1 ## Fgf9 1.039654e-09 1.0600988 0.163 0.054 4.148739e-05 1 ## Edem2 1.047110e-09 0.3432218 0.653 0.391 4.178494e-05 1 ## Pfdn2 1.055989e-09 0.3929707 0.841 0.570 4.213922e-05 1 ## Hps1 1.070998e-09 0.7108863 0.273 0.122 4.273818e-05 1 ## Dynlt2b 1.080433e-09 0.7973710 0.351 0.177 4.311468e-05 1 ## Sms1 1.084417e-09 0.3879869 0.759 0.502 4.327366e-05 1 ## Gab1 1.087769e-09 0.6303987 0.331 0.161 4.340744e-05 1 ## Nin 1.088697e-09 0.2528326 0.527 0.278 4.344446e-05 1 ## Adprh1 1.090326e-09 0.4863875 0.506 0.286 4.350946e-05 1 ## Bad 1.094057e-09 0.4579702 0.800 0.512 4.365835e-05 1 ## Zfp444 1.128884e-09 0.5004489 0.445 0.239 4.504810e-05 1 ## Sub11 1.128915e-09 0.3042034 0.943 0.651 4.504935e-05 1 ## Rab3ip 1.159064e-09 0.5123275 0.673 0.422 4.625244e-05 1 ## Sowahc1 1.160973e-09 0.2942281 0.759 0.481 4.632861e-05 1 ## Mea11 1.162503e-09 0.5318776 0.641 0.386 4.638970e-05 1 ## Surf1 1.174673e-09 0.5865339 0.514 0.290 4.687533e-05 1 ## Atp5me1 1.180285e-09 0.1684859 0.996 0.818 4.709928e-05 1 ## Fsd1l1 1.190345e-09 0.2100813 0.951 0.699 4.750071e-05 1 ## A030001D20Rik 1.227696e-09 2.1209163 0.073 0.013 4.899122e-05 1 ## B230354K17Rik 1.240390e-09 0.6521840 0.331 0.159 4.949776e-05 1 ## Cyp4v31 1.300735e-09 0.1085644 0.853 0.556 5.190583e-05 1 ## Med7 1.301901e-09 0.5238508 0.424 0.215 5.195235e-05 1 ## 2010320M18Rik 1.313968e-09 0.5536531 0.380 0.189 5.243391e-05 1 ## Asphd1 1.323453e-09 0.7921537 0.216 0.082 5.281240e-05 1 ## Samd9l 1.326014e-09 0.2288889 0.731 0.457 5.291460e-05 1 ## Gm46120 1.334940e-09 2.0172830 0.057 0.007 5.327080e-05 1 ## Igsf231 1.369256e-09 0.2532393 0.792 0.505 5.464016e-05 1 ## Tm2d21 1.369355e-09 0.3671660 0.792 0.509 5.464413e-05 1 ## Nudt13 1.372860e-09 0.3434265 0.457 0.244 5.478396e-05 1 ## Trp53i13 1.397013e-09 0.3693408 0.567 0.312 5.574782e-05 1 ## D330023K18Rik 1.409716e-09 0.9073378 0.253 0.108 5.625472e-05 1 ## Psma31 1.416179e-09 0.3138548 0.967 0.720 5.651264e-05 1 ## Tmem108 1.423682e-09 0.8145806 0.220 0.086 5.681201e-05 1 ## Gramd1a 1.434202e-09 0.5293599 0.290 0.133 5.723182e-05 1 ## Limch1 1.440656e-09 0.7591709 0.233 0.094 5.748938e-05 1 ## Got1 1.449814e-09 0.2866329 0.363 0.175 5.785481e-05 1 ## Arb2a1 1.452489e-09 0.2670399 0.812 0.531 5.796159e-05 1 ## Gm39271 1.466349e-09 2.4178224 0.065 0.010 5.851466e-05 1 ## Fam3a 1.466771e-09 0.4234183 0.478 0.259 5.853150e-05 1 ## Rtl61 1.491307e-09 0.5367544 0.339 0.156 5.951061e-05 1 ## Echdc2 1.522528e-09 0.4795863 0.396 0.208 6.075649e-05 1 ## Eci1 1.551025e-09 0.3583990 0.739 0.470 6.189364e-05 1 ## Smyd2 1.551627e-09 0.2409747 0.551 0.298 6.191768e-05 1 ## Smu1 1.558089e-09 0.4217205 0.608 0.358 6.217556e-05 1 ## Xylt1 1.561331e-09 1.9945766 0.073 0.013 6.230491e-05 1 ## Vps251 1.582244e-09 0.3546700 0.759 0.494 6.313945e-05 1 ## Ubac1 1.591862e-09 0.4510813 0.445 0.242 6.352324e-05 1 ## Cops8 1.618063e-09 0.3204283 0.547 0.306 6.456882e-05 1 ## Tceal1 1.643184e-09 0.7761613 0.278 0.119 6.557124e-05 1 ## Psmb2 1.670667e-09 0.2319644 0.914 0.600 6.666795e-05 1 ## Orai3 1.679788e-09 0.7303023 0.306 0.138 6.703195e-05 1 ## Dis3l2 1.704958e-09 0.6185321 0.343 0.164 6.803633e-05 1 ## Cul5 1.707928e-09 0.4124193 0.649 0.393 6.815486e-05 1 ## Pafah1b3 1.712243e-09 0.2676664 0.722 0.437 6.832706e-05 1 ## Rad21l 1.726961e-09 2.7060905 0.041 0.003 6.891437e-05 1 ## H4c4 1.732742e-09 0.2183866 0.645 0.386 6.914506e-05 1 ## Smim26 1.748160e-09 0.4884184 0.657 0.421 6.976031e-05 1 ## Sorbs2 1.824853e-09 0.1257421 0.543 0.298 7.282077e-05 1 ## Enpp5 1.840988e-09 0.5666764 0.412 0.209 7.346463e-05 1 ## Znrd21 1.842240e-09 0.5165657 0.473 0.256 7.351460e-05 1 ## Atp6v1c11 1.847798e-09 0.4182354 0.755 0.498 7.373637e-05 1 ## Bet1l 1.872202e-09 0.4406041 0.722 0.465 7.471023e-05 1 ## Rab211 1.888645e-09 0.4416908 0.678 0.432 7.536636e-05 1 ## Mfsd10 1.905088e-09 0.9381469 0.290 0.136 7.602255e-05 1 ## Kif3a 1.905831e-09 0.4270452 0.482 0.266 7.605217e-05 1 ## Efhd21 1.910767e-09 0.2871905 0.959 0.672 7.624915e-05 1 ## Alyref2 1.910975e-09 0.8588022 0.273 0.122 7.625747e-05 1 ## Zfp26 1.925225e-09 0.4033023 0.490 0.270 7.682611e-05 1 ## Pdia4 1.941126e-09 0.3392258 0.820 0.534 7.746064e-05 1 ## Pigbos1 1.964066e-09 0.4235961 0.735 0.439 7.837605e-05 1 ## Slc17a5 2.006393e-09 0.2658723 0.702 0.437 8.006513e-05 1 ## Arhgap11 2.044339e-09 0.3535020 0.669 0.405 8.157936e-05 1 ## Ablim11 2.080672e-09 0.3093645 0.865 0.607 8.302921e-05 1 ## Mtg1 2.081097e-09 1.0273683 0.220 0.090 8.304618e-05 1 ## Bcl10 2.084878e-09 0.3647242 0.645 0.398 8.319707e-05 1 ## Rnh11 2.097033e-09 0.3238720 0.882 0.613 8.368211e-05 1 ## Nfe2l21 2.103020e-09 0.3240378 0.857 0.634 8.392103e-05 1 ## Miga1 2.131977e-09 0.4179492 0.371 0.187 8.507653e-05 1 ## Degs1 2.136589e-09 0.7570398 0.400 0.215 8.526057e-05 1 ## Ndufv2 2.142999e-09 0.1755502 0.935 0.632 8.551639e-05 1 ## Fam120aos1 2.190811e-09 0.3924729 0.608 0.356 8.742433e-05 1 ## Trappc2 2.228984e-09 0.7156882 0.310 0.143 8.894759e-05 1 ## Serinc3 2.270148e-09 0.1945383 0.914 0.646 9.059025e-05 1 ## Cflar 2.282052e-09 0.3097785 0.763 0.490 9.106529e-05 1 ## Faap20 2.339988e-09 0.4888186 0.576 0.330 9.337721e-05 1 ## Os91 2.346482e-09 0.2563216 0.971 0.772 9.363637e-05 1 ## Commd61 2.355496e-09 0.3720295 0.804 0.531 9.399606e-05 1 ## Qtrt1 2.378414e-09 0.6707635 0.331 0.159 9.491060e-05 1 ## Letmd1 2.398004e-09 0.8792593 0.339 0.166 9.569235e-05 1 ## Rtl8a 2.477226e-09 1.0260410 0.176 0.061 9.885371e-05 1 ## Calm11 2.505080e-09 0.1773211 1.000 0.951 9.996521e-05 1 ## Gm44066 2.507779e-09 1.5763922 0.122 0.035 1.000729e-04 1 ## Atp1b3 2.538530e-09 0.4431311 0.653 0.400 1.013000e-04 1 ## Nedd41 2.575007e-09 0.2900647 0.980 0.802 1.027556e-04 1 ## Spa17 2.628019e-09 0.5870493 0.400 0.208 1.048711e-04 1 ## Scrn1 2.678932e-09 0.4211682 0.645 0.389 1.069028e-04 1 ## Rhobtb3 2.687857e-09 0.3761655 0.367 0.184 1.072589e-04 1 ## Vegfb 2.694193e-09 0.4799516 0.388 0.196 1.075118e-04 1 ## Amotl1 2.794922e-09 1.7299522 0.102 0.025 1.115314e-04 1 ## Lman1 2.870598e-09 0.4555265 0.771 0.510 1.145512e-04 1 ## Zfpl1 2.889543e-09 0.4660088 0.518 0.290 1.153072e-04 1 ## Ift27 2.916051e-09 0.6183737 0.400 0.203 1.163650e-04 1 ## Dnaja41 2.918006e-09 0.3440695 0.894 0.639 1.164430e-04 1 ## Lrba 2.924394e-09 0.2507734 0.841 0.562 1.166980e-04 1 ## Plekhj1 2.953971e-09 0.3230962 0.747 0.471 1.178782e-04 1 ## Tapbpl 2.954771e-09 0.6198160 0.522 0.317 1.179101e-04 1 ## Irf1 2.959854e-09 0.2320460 0.820 0.551 1.181130e-04 1 ## Sf3b5 3.046496e-09 0.1406581 0.759 0.450 1.215704e-04 1 ## Ppp1ca1 3.054838e-09 0.2797577 0.992 0.766 1.219033e-04 1 ## Carlr 3.065985e-09 4.2179572 0.024 0.000 1.223481e-04 1 ## Halr1 3.065985e-09 4.4555403 0.024 0.000 1.223481e-04 1 ## Atp8b3 3.065985e-09 4.4128660 0.024 0.000 1.223481e-04 1 ## Gm42060 3.065985e-09 4.2591057 0.024 0.000 1.223481e-04 1 ## Gm32144 3.065985e-09 4.4284801 0.024 0.000 1.223481e-04 1 ## Igsf8 3.068820e-09 0.4302547 0.498 0.279 1.224613e-04 1 ## Ptpn9 3.101455e-09 0.4431506 0.547 0.314 1.237636e-04 1 ## Smagp1 3.102585e-09 0.3694962 0.661 0.402 1.238087e-04 1 ## Slc25a11 3.113706e-09 0.3889871 0.808 0.518 1.242525e-04 1 ## Slc39a10 3.182411e-09 0.5447020 0.294 0.135 1.269941e-04 1 ## Atp8b2 3.227930e-09 1.1105257 0.143 0.045 1.288105e-04 1 ## Sdf21 3.230694e-09 0.3180509 0.739 0.479 1.289208e-04 1 ## Tpd52l1 3.233446e-09 0.7958529 0.351 0.173 1.290307e-04 1 ## Iscu 3.275027e-09 0.2025738 0.751 0.486 1.306900e-04 1 ## Pdia31 3.280975e-09 0.2434082 0.996 0.850 1.309273e-04 1 ## Zfp882 3.305432e-09 0.7524424 0.216 0.084 1.319033e-04 1 ## Gm34552 3.471827e-09 1.0189200 0.139 0.043 1.385433e-04 1 ## Fbxo3 3.500568e-09 0.2845343 0.771 0.498 1.396902e-04 1 ## Rtn31 3.502212e-09 0.3039029 0.963 0.741 1.397558e-04 1 ## Lyrm2 3.518448e-09 0.3570807 0.633 0.381 1.404037e-04 1 ## Cdkn2d 3.520935e-09 0.5788708 0.445 0.236 1.405029e-04 1 ## Hacd31 3.580337e-09 0.3367883 0.722 0.468 1.428733e-04 1 ## Tifa 3.601625e-09 0.3536380 0.449 0.238 1.437228e-04 1 ## Psme2 3.635418e-09 0.1591450 0.943 0.694 1.450713e-04 1 ## Zfand1 3.723739e-09 0.6837167 0.388 0.203 1.485958e-04 1 ## Slc43a21 3.845665e-09 0.1343072 0.861 0.554 1.534613e-04 1 ## Srsf91 3.978423e-09 0.4118108 0.792 0.521 1.587590e-04 1 ## Mat2b 4.004882e-09 0.4089888 0.714 0.460 1.598148e-04 1 ## Rab6b1 4.021128e-09 0.6003188 0.396 0.203 1.604631e-04 1 ## Mrpl22 4.033775e-09 0.4127124 0.498 0.270 1.609678e-04 1 ## Slc39a61 4.073410e-09 0.4975526 0.437 0.233 1.625494e-04 1 ## Snx21 4.133542e-09 0.9315412 0.261 0.116 1.649490e-04 1 ## Irgm1 4.182026e-09 0.9847456 0.261 0.119 1.668837e-04 1 ## Rtn4rl11 4.216400e-09 0.7117357 0.457 0.253 1.682554e-04 1 ## Lca5 4.271108e-09 1.1686127 0.135 0.041 1.704386e-04 1 ## Med31 4.302060e-09 0.6974470 0.441 0.242 1.716737e-04 1 ## Patz1 4.353453e-09 0.6950672 0.347 0.168 1.737246e-04 1 ## Taf101 4.417339e-09 0.2440910 0.963 0.694 1.762739e-04 1 ## Gm15520 4.451704e-09 3.9958185 0.033 0.001 1.776453e-04 1 ## Gm6329 4.509728e-09 2.3787149 0.061 0.009 1.799607e-04 1 ## Nt5c3b 4.558780e-09 0.4382226 0.269 0.120 1.819181e-04 1 ## Chrac1 4.605329e-09 0.4693934 0.624 0.374 1.837757e-04 1 ## Rapgef3os2 4.657383e-09 3.3528272 0.033 0.001 1.858529e-04 1 ## Tstd3 4.692483e-09 0.5920687 0.269 0.122 1.872535e-04 1 ## Chmp51 4.692589e-09 0.2392525 0.951 0.714 1.872578e-04 1 ## Spty2d1 4.789159e-09 0.3630246 0.555 0.328 1.911114e-04 1 ## Stat5b 4.815980e-09 0.5891665 0.384 0.199 1.921817e-04 1 ## Apbb11 4.825549e-09 0.4179624 0.465 0.249 1.925635e-04 1 ## Mrps121 4.830008e-09 0.3415828 0.820 0.522 1.927415e-04 1 ## Arpc21 4.957695e-09 0.2414824 0.996 0.867 1.978368e-04 1 ## Maml3 5.028156e-09 0.3120717 0.763 0.506 2.006486e-04 1 ## Gm33055 5.037112e-09 4.4325139 0.029 0.001 2.010059e-04 1 ## Vps36 5.045369e-09 0.4131721 0.714 0.451 2.013354e-04 1 ## Tmem151a 5.163916e-09 0.4089289 0.388 0.189 2.060661e-04 1 ## Arl15 5.234564e-09 0.6361377 0.253 0.110 2.088853e-04 1 ## Pacs1 5.245853e-09 0.7105236 0.212 0.084 2.093358e-04 1 ## Ank3 5.250742e-09 0.1047187 0.931 0.713 2.095309e-04 1 ## Herpud2 5.390313e-09 0.4277518 0.808 0.512 2.151004e-04 1 ## Txnl1 5.426895e-09 0.2851114 0.865 0.586 2.165602e-04 1 ## Surf4 5.463012e-09 0.3014084 0.943 0.697 2.180015e-04 1 ## Anp32b1 5.474742e-09 0.1960776 0.988 0.775 2.184696e-04 1 ## Kctd13 5.491259e-09 0.6293194 0.318 0.149 2.191287e-04 1 ## Cds2 5.521350e-09 0.2422363 0.686 0.425 2.203295e-04 1 ## 2810001G20Rik 5.555513e-09 0.7711742 0.286 0.132 2.216927e-04 1 ## Dnmt3b 5.618152e-09 0.9803817 0.176 0.062 2.241924e-04 1 ## Furin 5.619096e-09 0.4118837 0.665 0.412 2.242300e-04 1 ## Naa38 5.649481e-09 0.3737696 0.673 0.422 2.254425e-04 1 ## 2410022M11Rik 5.713093e-09 1.0117652 0.176 0.063 2.279810e-04 1 ## 4921524J17Rik 5.724436e-09 0.6457892 0.547 0.328 2.284336e-04 1 ## Mtch11 5.728449e-09 0.2443394 0.988 0.832 2.285938e-04 1 ## Tmem267 5.769509e-09 1.0353373 0.192 0.073 2.302323e-04 1 ## Iyd1 5.865172e-09 0.2307603 0.584 0.330 2.340497e-04 1 ## Hnrnph3 5.884368e-09 0.2707033 0.661 0.397 2.348157e-04 1 ## A930005H10Rik 5.914125e-09 0.8397021 0.282 0.133 2.360032e-04 1 ## Slc25a23 6.005853e-09 1.0204665 0.216 0.088 2.396636e-04 1 ## Ifit3 6.014931e-09 1.3580951 0.114 0.031 2.400258e-04 1 ## Gm54326 6.025417e-09 0.4321283 0.510 0.290 2.404443e-04 1 ## Ak1 6.072737e-09 1.0719658 0.163 0.055 2.423326e-04 1 ## Etfb1 6.107113e-09 0.2271467 0.943 0.711 2.437043e-04 1 ## Vcf11 6.169416e-09 0.3088125 0.906 0.648 2.461905e-04 1 ## Sec22a1 6.205188e-09 0.4711183 0.449 0.247 2.476180e-04 1 ## Eny2 6.219479e-09 0.2882259 0.878 0.600 2.481883e-04 1 ## Cul4a 6.325923e-09 0.2897561 0.563 0.329 2.524360e-04 1 ## Triap1 6.345726e-09 0.5130271 0.571 0.329 2.532262e-04 1 ## Txndc91 6.434179e-09 0.4364436 0.796 0.526 2.567559e-04 1 ## Snrpb 6.528929e-09 0.1755978 0.890 0.595 2.605369e-04 1 ## 1500026H17Rik 6.604024e-09 1.0211239 0.245 0.106 2.635336e-04 1 ## Tspan311 6.636845e-09 0.2453917 0.865 0.604 2.648433e-04 1 ## Uckl1 6.693119e-09 0.5012245 0.539 0.319 2.670889e-04 1 ## Ywhab1 6.707255e-09 0.2521909 0.988 0.825 2.676530e-04 1 ## Slc30a7 6.794903e-09 0.4050212 0.637 0.398 2.711506e-04 1 ## Rpl34-ps1 6.925811e-09 0.2436145 0.612 0.348 2.763745e-04 1 ## Srp91 6.995804e-09 0.2564220 0.922 0.651 2.791675e-04 1 ## Dnajc1 7.039110e-09 0.3942316 0.641 0.387 2.808957e-04 1 ## Rnf142 7.081417e-09 0.3845600 0.714 0.463 2.825840e-04 1 ## Rfx5 7.262427e-09 0.5248505 0.265 0.117 2.898071e-04 1 ## Gm10642 7.314461e-09 1.3524495 0.127 0.037 2.918836e-04 1 ## Hsd17b4 7.334786e-09 0.1186184 0.592 0.335 2.926946e-04 1 ## Tnks21 7.457353e-09 0.2674792 0.890 0.674 2.975857e-04 1 ## Cspg4b 7.465776e-09 2.8106909 0.049 0.006 2.979218e-04 1 ## A930028N01Rik 7.495593e-09 1.0451909 0.192 0.073 2.991117e-04 1 ## Kank3 7.505936e-09 1.9604878 0.078 0.015 2.995244e-04 1 ## Mydgf1 7.585360e-09 0.3552739 0.906 0.634 3.026938e-04 1 ## Amacr 7.611270e-09 0.7285379 0.318 0.157 3.037277e-04 1 ## Tcta1 7.623268e-09 0.4431676 0.571 0.339 3.042065e-04 1 ## Syt17 7.640282e-09 1.6892138 0.086 0.019 3.048854e-04 1 ## Pkm1 7.718852e-09 0.1669404 1.000 0.915 3.080208e-04 1 ## Slc39a13 7.949850e-09 0.6883835 0.286 0.131 3.172388e-04 1 ## Eif1a 7.985046e-09 0.3280804 0.784 0.488 3.186433e-04 1 ## Gm52284 8.356413e-09 2.4750126 0.049 0.006 3.334626e-04 1 ## Lcmt1 8.383425e-09 0.6514446 0.424 0.228 3.345406e-04 1 ## Miat 8.389628e-09 0.3398967 0.278 0.126 3.347881e-04 1 ## Grk21 8.421702e-09 0.2380940 0.698 0.453 3.360680e-04 1 ## Gsdma1 8.446941e-09 0.1763120 0.845 0.518 3.370752e-04 1 ## Naa20 8.452704e-09 0.4270146 0.682 0.423 3.373052e-04 1 ## Abcc1 8.482136e-09 0.4833133 0.396 0.212 3.384796e-04 1 ## Ppp1r7 8.494164e-09 0.4772300 0.543 0.317 3.389596e-04 1 ## Btbd10 8.538781e-09 0.4612233 0.384 0.196 3.407400e-04 1 ## Chic2 8.750349e-09 0.4057521 0.682 0.431 3.491827e-04 1 ## Tmem101 9.009263e-09 0.5171057 0.449 0.242 3.595146e-04 1 ## Oma1 9.090002e-09 0.5414822 0.396 0.217 3.627365e-04 1 ## Tpp11 9.135211e-09 0.2507625 0.657 0.400 3.645406e-04 1 ## Copb21 9.139074e-09 0.2658184 0.955 0.707 3.646948e-04 1 ## Zwint1 9.139387e-09 0.2555133 0.771 0.495 3.647072e-04 1 ## Gm33989 9.214624e-09 0.2576809 0.555 0.319 3.677096e-04 1 ## Angpt2 9.490696e-09 2.0228820 0.069 0.013 3.787262e-04 1 ## Rnaseh2c 9.532904e-09 0.3482663 0.580 0.331 3.804105e-04 1 ## Sergef 9.734194e-09 0.3818104 0.347 0.170 3.884430e-04 1 ## Ptma1 9.776835e-09 0.1004655 1.000 0.937 3.901446e-04 1 ## Adi11 9.998967e-09 0.2789432 0.747 0.469 3.990088e-04 1 ## Naxe 1.000179e-08 0.4389393 0.629 0.393 3.991214e-04 1 ## Nedd4l 1.004818e-08 0.3572315 0.678 0.417 4.009726e-04 1 ## Irak1 1.007219e-08 0.2338151 0.735 0.495 4.019307e-04 1 ## Map9 1.007930e-08 0.2212489 0.592 0.331 4.022145e-04 1 ## Taf13 1.009731e-08 0.6861685 0.331 0.166 4.029332e-04 1 ## Grik5 1.016950e-08 0.2675288 0.490 0.271 4.058139e-04 1 ## Pea15a1 1.023656e-08 0.5396007 0.339 0.164 4.084899e-04 1 ## Snrnp35 1.029846e-08 0.5940679 0.318 0.156 4.109599e-04 1 ## Phf10 1.036181e-08 0.4957805 0.518 0.299 4.134880e-04 1 ## Dtymk 1.039025e-08 0.1140237 0.433 0.222 4.146230e-04 1 ## Drap11 1.093146e-08 0.2965968 0.922 0.684 4.362198e-04 1 ## Ski 1.123985e-08 0.3020408 0.620 0.373 4.485264e-04 1 ## Jph4 1.137408e-08 1.0129506 0.224 0.094 4.538826e-04 1 ## Xkr4 1.162073e-08 1.2619033 0.098 0.024 4.637254e-04 1 ## Nelfb 1.165371e-08 0.4724309 0.527 0.314 4.650413e-04 1 ## Zfand6 1.172994e-08 0.2391692 0.927 0.665 4.680834e-04 1 ## Nuak21 1.173589e-08 0.2761341 0.829 0.546 4.683207e-04 1 ## Psmd121 1.208666e-08 0.2436106 0.853 0.585 4.823183e-04 1 ## Rtcb 1.219273e-08 0.2981623 0.714 0.428 4.865508e-04 1 ## Rpp38 1.236934e-08 0.6168192 0.306 0.150 4.935985e-04 1 ## Snrpd3 1.268470e-08 0.1222290 0.804 0.496 5.061830e-04 1 ## Hprt11 1.273712e-08 0.2868803 0.788 0.541 5.082749e-04 1 ## Vamp4 1.287375e-08 0.2585351 0.604 0.349 5.137269e-04 1 ## Echdc3 1.293675e-08 2.2207015 0.078 0.016 5.162409e-04 1 ## Atxn2 1.308465e-08 0.3285415 0.641 0.398 5.221429e-04 1 ## Yipf31 1.321587e-08 0.3259832 0.861 0.604 5.273795e-04 1 ## Gm38563 1.330123e-08 1.9356780 0.078 0.016 5.307856e-04 1 ## Cep19 1.349129e-08 0.3618455 0.453 0.254 5.383700e-04 1 ## Eef2 1.364065e-08 0.1961882 0.996 0.918 5.443303e-04 1 ## Gm4013 1.365730e-08 0.8379687 0.408 0.228 5.449944e-04 1 ## Tmem179b 1.366058e-08 0.2863236 0.563 0.337 5.451254e-04 1 ## Usp14 1.378086e-08 0.1999463 0.678 0.400 5.499251e-04 1 ## Rab42 1.384762e-08 1.6602404 0.057 0.008 5.525894e-04 1 ## Phpt11 1.405658e-08 0.3313970 0.714 0.423 5.609280e-04 1 ## Fam149a1 1.410715e-08 0.6456541 0.290 0.133 5.629457e-04 1 ## Nectin2 1.452859e-08 0.3349267 0.665 0.410 5.797635e-04 1 ## Klhl7 1.467487e-08 0.6334124 0.624 0.369 5.856009e-04 1 ## Trim27 1.472416e-08 0.5058675 0.494 0.283 5.875677e-04 1 ## Ccdc136 1.481395e-08 0.5611632 0.188 0.070 5.911508e-04 1 ## Cd83 1.486942e-08 1.2873053 0.159 0.058 5.933640e-04 1 ## Zfp974 1.493592e-08 0.5679613 0.245 0.105 5.960179e-04 1 ## Cct8 1.498132e-08 0.2827218 0.935 0.679 5.978295e-04 1 ## Ndufb61 1.503921e-08 0.2330719 0.878 0.595 6.001395e-04 1 ## Ccdc88a 1.518882e-08 0.4419336 0.269 0.122 6.061099e-04 1 ## Fchsd2 1.542164e-08 0.3041529 0.576 0.344 6.154006e-04 1 ## Eef1a2 1.544657e-08 1.2792379 0.061 0.010 6.163954e-04 1 ## Anapc10 1.556417e-08 0.6653522 0.351 0.179 6.210883e-04 1 ## Mrpl34 1.560410e-08 0.2144762 0.853 0.559 6.226818e-04 1 ## Spon2 1.561818e-08 1.3819744 0.061 0.010 6.232436e-04 1 ## Cand11 1.565224e-08 0.2777290 0.714 0.471 6.246028e-04 1 ## B4galt2 1.580372e-08 2.2972244 0.078 0.016 6.306476e-04 1 ## Gm52310 1.580372e-08 1.8098361 0.078 0.016 6.306476e-04 1 ## Map1lc3a1 1.581070e-08 0.2606605 0.804 0.540 6.309261e-04 1 ## Pik3c2a 1.601075e-08 0.3331178 0.727 0.502 6.389090e-04 1 ## Ufm11 1.606820e-08 0.3675433 0.845 0.593 6.412013e-04 1 ## Diaph3 1.621681e-08 0.5675566 0.216 0.091 6.471316e-04 1 ## Enpp41 1.632954e-08 0.5266172 0.396 0.210 6.516301e-04 1 ## M6pr 1.633669e-08 0.3997373 0.755 0.497 6.519157e-04 1 ## Cenpp 1.640005e-08 0.1787666 0.220 0.094 6.544441e-04 1 ## Ptger3 1.667021e-08 0.5456760 0.167 0.059 6.652248e-04 1 ## Nmt2 1.667711e-08 0.2706825 0.629 0.375 6.654999e-04 1 ## Loricrin 1.669495e-08 1.1075801 0.139 0.045 6.662122e-04 1 ## Trim3 1.676339e-08 0.4242337 0.314 0.148 6.689431e-04 1 ## Lasp1 1.685882e-08 0.1519733 0.918 0.632 6.727514e-04 1 ## Trappc5 1.688913e-08 0.2953265 0.665 0.403 6.739609e-04 1 ## Psmd81 1.724605e-08 0.3286274 0.914 0.637 6.882035e-04 1 ## Bhlhb9 1.752454e-08 0.4765739 0.327 0.160 6.993167e-04 1 ## Gm26782 1.784459e-08 0.3407620 0.465 0.256 7.120883e-04 1 ## Gnb51 1.799325e-08 0.4876857 0.404 0.211 7.180207e-04 1 ## Ccs 1.819783e-08 0.4181476 0.547 0.317 7.261844e-04 1 ## Rpl27 1.831836e-08 0.1093366 1.000 0.862 7.309943e-04 1 ## Snx14 1.835819e-08 0.4574206 0.522 0.309 7.325836e-04 1 ## Zfp580 1.838045e-08 1.3482008 0.143 0.048 7.334718e-04 1 ## Eef1d 1.841731e-08 0.1796083 0.959 0.678 7.349429e-04 1 ## Wbp12 1.872008e-08 0.2888530 0.673 0.421 7.470250e-04 1 ## Pik3ip1 1.880432e-08 0.9531270 0.167 0.061 7.503864e-04 1 ## Tle2 1.918578e-08 0.1601295 0.576 0.335 7.656087e-04 1 ## Gtf2a21 1.929303e-08 0.2834017 0.853 0.612 7.698884e-04 1 ## Pop51 1.957661e-08 0.3162237 0.845 0.570 7.812046e-04 1 ## Leo1 1.975378e-08 0.2880207 0.502 0.275 7.882747e-04 1 ## 2510002D24Rik 2.013223e-08 0.2755306 0.718 0.452 8.033768e-04 1 ## Epb41l4b1 2.060355e-08 0.1247754 0.988 0.795 8.221845e-04 1 ## Mrpl28 2.103155e-08 0.2217690 0.796 0.495 8.392638e-04 1 ## Polr2i1 2.108294e-08 0.3566586 0.698 0.430 8.413148e-04 1 ## Dhx30 2.122257e-08 0.3279681 0.461 0.257 8.468867e-04 1 ## Gm38661 2.126422e-08 2.5308349 0.045 0.005 8.485488e-04 1 ## Yif1b 2.130364e-08 0.5706726 0.531 0.310 8.501216e-04 1 ## Camk2n21 2.152501e-08 0.3319927 0.698 0.430 8.589553e-04 1 ## Fdft11 2.157794e-08 0.2374325 0.514 0.298 8.610675e-04 1 ## Lrrc31 2.201346e-08 1.1392403 0.212 0.091 8.784471e-04 1 ## Wdr25 2.208441e-08 1.6593112 0.090 0.022 8.812782e-04 1 ## Prkca 2.224812e-08 0.1930474 0.886 0.623 8.878114e-04 1 ## Adam101 2.226812e-08 0.3346891 0.788 0.522 8.886092e-04 1 ## Itgb3bp 2.248021e-08 0.5610820 0.343 0.176 8.970729e-04 1 ## Rmdn3 2.256038e-08 0.4012746 0.527 0.312 9.002721e-04 1 ## Zfp442 2.304228e-08 0.5103320 0.327 0.165 9.195021e-04 1 ## Ufsp2 2.373269e-08 0.3406454 0.735 0.466 9.470530e-04 1 ## Kcnj16 2.379513e-08 3.0644646 0.037 0.003 9.495447e-04 1 ## Sbsn 2.391345e-08 0.6479546 0.265 0.120 9.542660e-04 1 ## Vps281 2.395481e-08 0.3166846 0.914 0.651 9.559167e-04 1 ## Gm16548 2.418417e-08 2.6178595 0.037 0.003 9.650692e-04 1 ## Dctn21 2.424495e-08 0.2597007 0.890 0.634 9.674949e-04 1 ## Adgrl1 2.437565e-08 0.3845476 0.433 0.235 9.727104e-04 1 ## Mrpl301 2.458791e-08 0.2193188 0.841 0.556 9.811806e-04 1 ## Vsig10 2.514063e-08 0.3586089 0.539 0.321 1.003237e-03 1 ## Lurap1l1 2.517515e-08 0.3559606 0.559 0.335 1.004614e-03 1 ## Taf11 2.538143e-08 0.5229500 0.420 0.232 1.012846e-03 1 ## Kmt5a1 2.561229e-08 0.2717881 0.984 0.755 1.022058e-03 1 ## Ppa2 2.567753e-08 0.4895415 0.563 0.344 1.024662e-03 1 ## Zfp763 2.591090e-08 1.1059790 0.176 0.067 1.033974e-03 1 ## Gcsh 2.622135e-08 0.5744810 0.347 0.175 1.046363e-03 1 ## Atp6v0a11 2.656382e-08 0.2196831 0.751 0.483 1.060029e-03 1 ## Daam11 2.713623e-08 0.3878757 0.608 0.375 1.082871e-03 1 ## Msra 2.714710e-08 0.3385891 0.506 0.284 1.083305e-03 1 ## Rhoq 2.719128e-08 0.4674643 0.314 0.161 1.085068e-03 1 ## Nop16 2.727459e-08 0.6598987 0.290 0.141 1.088392e-03 1 ## Gm4035 2.770407e-08 3.0672104 0.049 0.006 1.105531e-03 1 ## Taf9b 2.784308e-08 0.7693659 0.224 0.095 1.111078e-03 1 ## Mthfsd 2.784593e-08 0.4778784 0.384 0.201 1.111192e-03 1 ## Nanp 2.785482e-08 0.3737718 0.359 0.187 1.111547e-03 1 ## 1700108F19Rik 2.810848e-08 3.0150464 0.057 0.009 1.121669e-03 1 ## Alkbh7 2.836814e-08 0.6447257 0.420 0.231 1.132031e-03 1 ## Kcnk11 2.864944e-08 0.2270228 0.816 0.494 1.143256e-03 1 ## Tmem1641 2.887326e-08 0.5159378 0.486 0.278 1.152187e-03 1 ## Mrps341 2.902058e-08 0.2962420 0.714 0.451 1.158066e-03 1 ## Mrpl231 2.914312e-08 0.1985727 0.739 0.463 1.162956e-03 1 ## Plxna3 2.921900e-08 0.5422197 0.176 0.066 1.165984e-03 1 ## Yeats41 2.924853e-08 0.2416257 0.755 0.484 1.167162e-03 1 ## Ramac1 2.986315e-08 0.3187143 0.788 0.514 1.191689e-03 1 ## C920006O11Rik 2.987748e-08 0.5210291 0.131 0.042 1.192261e-03 1 ## Chil6 2.995163e-08 2.2524118 0.065 0.012 1.195220e-03 1 ## Gm42094 3.008385e-08 2.2055298 0.090 0.022 1.200496e-03 1 ## Kcnb1 3.090603e-08 0.2914638 0.494 0.276 1.233305e-03 1 ## Bbox1 3.169023e-08 2.0393822 0.078 0.017 1.264599e-03 1 ## Hook3 3.180578e-08 0.2085572 0.759 0.496 1.269210e-03 1 ## Tnnt11 3.181213e-08 0.6895876 0.302 0.146 1.269463e-03 1 ## Tnfaip8 3.225725e-08 1.0940627 0.122 0.038 1.287225e-03 1 ## Nae1 3.244650e-08 0.2986920 0.551 0.335 1.294778e-03 1 ## Abca1 3.281052e-08 0.7685080 0.184 0.071 1.309304e-03 1 ## Parp2 3.285029e-08 0.3773509 0.482 0.269 1.310891e-03 1 ## Grpel2 3.344510e-08 0.2908400 0.718 0.461 1.334627e-03 1 ## Rabep11 3.350875e-08 0.2345202 0.869 0.623 1.337167e-03 1 ## Tsfm 3.397289e-08 0.4533976 0.400 0.208 1.355688e-03 1 ## Trpc1 3.410557e-08 0.4781802 0.310 0.147 1.360983e-03 1 ## Sel1l1 3.420112e-08 0.3086620 0.841 0.622 1.364796e-03 1 ## Gale1 3.454104e-08 0.1177211 0.763 0.472 1.378360e-03 1 ## 5430400D12Rik 3.540513e-08 0.9581985 0.155 0.056 1.412842e-03 1 ## Ogt 3.547220e-08 0.2866191 0.714 0.454 1.415518e-03 1 ## Foxk1 3.552951e-08 0.4279139 0.298 0.145 1.417805e-03 1 ## Faah 3.608460e-08 0.2329765 0.714 0.441 1.439956e-03 1 ## Fyn1 3.676234e-08 0.2645080 0.461 0.256 1.467001e-03 1 ## 2700062C07Rik 3.686322e-08 0.6084019 0.335 0.174 1.471027e-03 1 ## Nicn1 3.689334e-08 0.5929497 0.343 0.176 1.472229e-03 1 ## Ap3s2 3.689754e-08 0.4894340 0.510 0.297 1.472396e-03 1 ## Ces2b 3.714275e-08 1.6027417 0.053 0.008 1.482182e-03 1 ## Dtd2 3.794852e-08 0.5251461 0.376 0.200 1.514336e-03 1 ## Rrp11 3.816504e-08 0.2591322 0.943 0.687 1.522976e-03 1 ## Agbl3 3.820981e-08 1.0081166 0.135 0.044 1.524762e-03 1 ## Arpc51 3.836172e-08 0.2204335 0.988 0.827 1.530824e-03 1 ## Cerkl 3.850999e-08 0.1421038 0.535 0.300 1.536741e-03 1 ## Zfp707 3.882968e-08 0.5392841 0.290 0.138 1.549498e-03 1 ## Map3k3 3.891558e-08 0.3977261 0.563 0.344 1.552926e-03 1 ## Noc2l 3.901446e-08 0.2722258 0.429 0.224 1.556872e-03 1 ## Gata4 3.917006e-08 0.2781137 0.584 0.354 1.563081e-03 1 ## Ppid 3.938922e-08 0.2455643 0.588 0.336 1.571827e-03 1 ## Crybg3 3.961657e-08 0.2723771 0.282 0.133 1.580899e-03 1 ## Tmc41 3.965262e-08 0.2238257 0.976 0.797 1.582338e-03 1 ## Add1 4.075986e-08 0.2289335 0.702 0.428 1.626522e-03 1 ## Ankrd46 4.136924e-08 0.4314866 0.424 0.231 1.650839e-03 1 ## Slc22a181 4.142661e-08 0.2774573 0.727 0.460 1.653129e-03 1 ## Lsm4 4.178241e-08 0.1407123 0.833 0.550 1.667327e-03 1 ## Kit 4.208409e-08 0.6446635 0.282 0.137 1.679366e-03 1 ## Pomgnt11 4.217184e-08 0.3663386 0.461 0.252 1.682867e-03 1 ## Trmt1121 4.228873e-08 0.3612135 0.735 0.474 1.687532e-03 1 ## Pkig 4.271876e-08 0.2749340 0.731 0.452 1.704692e-03 1 ## 1700007L15Rik 4.277284e-08 1.4852359 0.147 0.052 1.706850e-03 1 ## Lurap1 4.282226e-08 2.0702640 0.057 0.009 1.708822e-03 1 ## Cyld 4.329114e-08 0.3957863 0.612 0.371 1.727533e-03 1 ## Ulk2 4.519715e-08 0.1315877 0.437 0.240 1.803592e-03 1 ## Cnrip1 4.526328e-08 3.1496543 0.033 0.002 1.806231e-03 1 ## Dipk1a 4.604147e-08 1.5137103 0.127 0.041 1.837285e-03 1 ## 5730455P16Rik 4.626989e-08 0.3508917 0.408 0.227 1.846400e-03 1 ## Rsph91 4.637988e-08 0.4942071 0.408 0.215 1.850789e-03 1 ## Gm34939 4.653689e-08 1.1793807 0.122 0.038 1.857055e-03 1 ## Abhd8 4.682340e-08 0.8584207 0.245 0.110 1.868488e-03 1 ## Wdfy4 4.685475e-08 2.1728013 0.033 0.002 1.869739e-03 1 ## Gm15860 4.752850e-08 1.1592137 0.188 0.077 1.896625e-03 1 ## Gm13375 4.801340e-08 0.9202034 0.151 0.054 1.915975e-03 1 ## Cep83os 4.864067e-08 1.2028397 0.163 0.061 1.941006e-03 1 ## AI837181 4.912421e-08 0.4118144 0.384 0.210 1.960302e-03 1 ## Sypl1 4.954012e-08 0.2559798 0.890 0.615 1.976899e-03 1 ## Lrp111 5.075273e-08 0.2755465 0.710 0.451 2.025288e-03 1 ## Naga1 5.137198e-08 0.2868674 0.645 0.422 2.049999e-03 1 ## Lpp1 5.138197e-08 0.2696086 0.927 0.727 2.050398e-03 1 ## Cacna1h1 5.150557e-08 0.1749874 0.678 0.408 2.055330e-03 1 ## Hikeshi 5.194084e-08 0.3762211 0.571 0.331 2.072699e-03 1 ## Ipp 5.226325e-08 0.4430876 0.412 0.229 2.085565e-03 1 ## Fkbp4 5.280604e-08 0.2017238 0.959 0.719 2.107225e-03 1 ## Gm34220 5.342114e-08 0.4703779 0.318 0.162 2.131771e-03 1 ## Timm17b 5.395183e-08 0.3072520 0.567 0.347 2.152948e-03 1 ## Pdia5 5.423955e-08 0.6671413 0.314 0.158 2.164429e-03 1 ## Zfp560 5.499681e-08 0.4237407 0.335 0.175 2.194648e-03 1 ## Ptgr21 5.555786e-08 0.3295956 0.645 0.410 2.217036e-03 1 ## Arpc5l1 5.571639e-08 0.3358350 0.865 0.594 2.223363e-03 1 ## Gm51482 5.637066e-08 2.7171050 0.041 0.004 2.249471e-03 1 ## Atp5mc1 5.658710e-08 0.1473283 1.000 0.817 2.258108e-03 1 ## Tmtc1 5.660623e-08 0.7309365 0.163 0.059 2.258872e-03 1 ## Cisd21 5.685112e-08 0.2463227 0.804 0.575 2.268644e-03 1 ## Ptprj 5.692082e-08 0.4836481 0.371 0.203 2.271425e-03 1 ## Rwdd2a 5.708295e-08 0.9745547 0.196 0.079 2.277895e-03 1 ## Akr1a11 5.767996e-08 0.2472103 0.996 0.839 2.301719e-03 1 ## Tbc1d8b 5.831393e-08 0.4471573 0.461 0.265 2.327017e-03 1 ## Cep15 5.834365e-08 0.5310809 0.527 0.325 2.328204e-03 1 ## Ndufs41 5.895035e-08 0.2516281 0.939 0.677 2.352414e-03 1 ## Yipf61 5.963881e-08 0.3835039 0.612 0.395 2.379887e-03 1 ## Snx5 6.001516e-08 0.3353960 0.722 0.463 2.394905e-03 1 ## Adcy61 6.015845e-08 0.2344577 0.759 0.494 2.400623e-03 1 ## Mrpl40 6.080479e-08 0.5233710 0.494 0.294 2.426415e-03 1 ## Ppp1cc1 6.129000e-08 0.3512849 0.596 0.363 2.445777e-03 1 ## A230070E04Rik 6.142496e-08 1.4457476 0.114 0.035 2.451163e-03 1 ## Trappc41 6.179027e-08 0.3541268 0.739 0.490 2.465741e-03 1 ## Hadhb 6.183979e-08 0.2070274 0.873 0.631 2.467717e-03 1 ## Pgam11 6.186258e-08 0.1577650 0.931 0.683 2.468626e-03 1 ## 1700054M17Rik 6.310208e-08 4.6121749 0.020 0.000 2.518089e-03 1 ## Gm37548 6.310208e-08 4.0538435 0.020 0.000 2.518089e-03 1 ## Sohlh2 6.310208e-08 4.0949498 0.020 0.000 2.518089e-03 1 ## Gm833 6.310208e-08 4.2186484 0.020 0.000 2.518089e-03 1 ## Gm46906 6.310208e-08 4.4535149 0.020 0.000 2.518089e-03 1 ## Gm46483 6.310208e-08 4.4195372 0.020 0.000 2.518089e-03 1 ## Tcp10c 6.310208e-08 4.3187194 0.020 0.000 2.518089e-03 1 ## Ube2g2 6.320619e-08 0.1392493 0.482 0.270 2.522243e-03 1 ## Lad1 6.388837e-08 0.1081565 0.592 0.352 2.549466e-03 1 ## Cdk41 6.452132e-08 0.1392973 0.743 0.462 2.574723e-03 1 ## Ip6k21 6.500409e-08 0.2825561 0.686 0.438 2.593988e-03 1 ## Hmox21 6.501754e-08 0.4042780 0.567 0.334 2.594525e-03 1 ## Hax11 6.532199e-08 0.3552645 0.514 0.305 2.606674e-03 1 ## Smim71 6.591480e-08 0.2531728 0.914 0.632 2.630330e-03 1 ## Zbtb14 6.652719e-08 0.9005906 0.241 0.111 2.654767e-03 1 ## Saysd1 6.673675e-08 0.4038540 0.535 0.317 2.663130e-03 1 ## Tstd1 6.729499e-08 0.4216130 0.261 0.122 2.685406e-03 1 ## Tex2611 6.811043e-08 0.1858209 0.780 0.484 2.717947e-03 1 ## Ube2d11 6.817163e-08 0.4808399 0.478 0.270 2.720389e-03 1 ## Stoml2 6.907860e-08 0.2884310 0.514 0.296 2.756582e-03 1 ## Mitd11 6.922100e-08 0.4175143 0.551 0.333 2.762264e-03 1 ## Ccdc92b 6.966258e-08 0.3114911 0.433 0.231 2.779885e-03 1 ## Dpcd 7.161176e-08 0.3677867 0.539 0.326 2.857667e-03 1 ## Hadh 7.191030e-08 0.1376135 0.959 0.727 2.869581e-03 1 ## Lrrcc1 7.232963e-08 0.2968480 0.371 0.196 2.886314e-03 1 ## Tgm5 7.250295e-08 1.3144851 0.114 0.035 2.893230e-03 1 ## Ftx 7.251674e-08 0.5879010 0.184 0.073 2.893781e-03 1 ## 2310001H17Rik 7.377146e-08 0.4138431 0.527 0.307 2.943850e-03 1 ## Zfp82 7.492602e-08 0.5699075 0.147 0.051 2.989923e-03 1 ## Slc7a6 7.527959e-08 0.3556969 0.282 0.133 3.004032e-03 1 ## Areg 7.561231e-08 1.3963440 0.143 0.051 3.017309e-03 1 ## Slc26a2 7.594778e-08 0.1382047 0.841 0.596 3.030696e-03 1 ## Fam171a2 7.604166e-08 1.8144944 0.090 0.023 3.034442e-03 1 ## Tmsb15b1 7.621159e-08 1.0361292 0.249 0.118 3.041223e-03 1 ## Eif3m1 7.711566e-08 0.2060867 0.833 0.546 3.077300e-03 1 ## Akirin2 7.774591e-08 0.2249352 0.739 0.464 3.102451e-03 1 ## Trmt10b 7.797764e-08 0.4542438 0.290 0.142 3.111698e-03 1 ## Chchd21 7.810109e-08 0.1423737 1.000 0.918 3.116624e-03 1 ## Zdhhc12 7.827967e-08 0.3401751 0.465 0.266 3.123750e-03 1 ## Dcps 7.850492e-08 1.0018607 0.265 0.128 3.132739e-03 1 ## Kifap3 7.869024e-08 0.2994753 0.563 0.338 3.140134e-03 1 ## Tdrd5 7.896321e-08 3.0403486 0.045 0.006 3.151027e-03 1 ## 2310030G06Rik 7.989553e-08 0.3669448 0.535 0.326 3.188231e-03 1 ## Klhl36 8.027750e-08 0.9669423 0.167 0.066 3.203474e-03 1 ## Ncoa7 8.093027e-08 0.2892153 0.592 0.360 3.229522e-03 1 ## Nipsnap11 8.132969e-08 0.2909808 0.735 0.484 3.245461e-03 1 ## Tmem132a 8.218850e-08 1.0543414 0.147 0.053 3.279732e-03 1 ## Keap11 8.277668e-08 0.3844417 0.678 0.442 3.303203e-03 1 ## Supt20 8.365953e-08 0.2621443 0.571 0.338 3.338433e-03 1 ## Sdsl 8.401820e-08 0.3978572 0.359 0.182 3.352746e-03 1 ## Zfpm11 8.402653e-08 0.1980353 0.922 0.688 3.353079e-03 1 ## Tsr11 8.403545e-08 0.3951786 0.384 0.203 3.353435e-03 1 ## Gm33682 8.437580e-08 0.8158730 0.098 0.027 3.367016e-03 1 ## Arhgap32 8.447458e-08 0.2101873 0.706 0.463 3.370958e-03 1 ## Cers4 8.469597e-08 0.4887751 0.465 0.273 3.379793e-03 1 ## Sac3d1 8.512614e-08 0.4566224 0.433 0.248 3.396959e-03 1 ## Chmp2a1 8.523142e-08 0.2173016 0.984 0.793 3.401160e-03 1 ## 1110004F10Rik1 8.579887e-08 0.3247252 0.865 0.614 3.423804e-03 1 ## Mia2 8.665928e-08 0.1876120 0.943 0.737 3.458139e-03 1 ## Pafah1b11 8.666284e-08 0.2245942 0.980 0.790 3.458281e-03 1 ## Plp2 8.685492e-08 0.2575063 0.739 0.453 3.465945e-03 1 ## Med11 8.730280e-08 0.4488447 0.359 0.194 3.483818e-03 1 ## Dda11 8.996959e-08 0.3424503 0.686 0.447 3.590236e-03 1 ## Diablo1 9.079178e-08 0.4202010 0.580 0.358 3.623046e-03 1 ## Sncg 9.333833e-08 0.2324000 0.078 0.017 3.724666e-03 1 ## Poglut1 9.383346e-08 0.5368689 0.302 0.152 3.744424e-03 1 ## Sec11c 9.434821e-08 0.2526274 0.837 0.567 3.764965e-03 1 ## Muc20 9.440095e-08 1.2774100 0.061 0.011 3.767070e-03 1 ## Prl6a1 9.445035e-08 4.1027335 0.024 0.001 3.769041e-03 1 ## Armc1 9.552082e-08 0.2652401 0.576 0.349 3.811758e-03 1 ## Gm46650 9.626132e-08 1.8691057 0.045 0.006 3.841308e-03 1 ## Cd46 9.635239e-08 1.4622730 0.106 0.031 3.844942e-03 1 ## Tram1l1 9.715096e-08 3.8404771 0.024 0.001 3.876809e-03 1 ## Gpr158 9.852914e-08 3.2510666 0.024 0.001 3.931805e-03 1 ## Mageh1 9.856289e-08 0.1895669 0.567 0.326 3.933152e-03 1 ## Gucd1 1.004499e-07 0.3635000 0.535 0.327 4.008453e-03 1 ## Ophn1 1.004833e-07 0.8115208 0.212 0.093 4.009785e-03 1 ## Slc39a7 1.013214e-07 0.2788378 0.710 0.458 4.043230e-03 1 ## Ctf1 1.017402e-07 1.3092077 0.143 0.051 4.059943e-03 1 ## Dexi 1.021678e-07 0.5786651 0.261 0.126 4.077008e-03 1 ## Zmpste241 1.027217e-07 0.3619100 0.588 0.360 4.099109e-03 1 ## Slc35d1 1.029734e-07 0.3747440 0.506 0.306 4.109154e-03 1 ## Tsix 1.036294e-07 0.2244370 0.429 0.236 4.135332e-03 1 ## Pcgf2 1.044986e-07 0.4094314 0.351 0.180 4.170016e-03 1 ## Klhdc10 1.051208e-07 0.2609413 0.563 0.348 4.194845e-03 1 ## Dock9 1.056412e-07 0.1599130 0.433 0.235 4.215612e-03 1 ## Tspan3 1.060211e-07 0.2221781 0.857 0.580 4.230771e-03 1 ## Stard101 1.079316e-07 0.1993914 0.996 0.863 4.307012e-03 1 ## Cldnd1 1.082503e-07 0.2525562 0.669 0.425 4.319727e-03 1 ## Ro601 1.083465e-07 0.3353520 0.665 0.438 4.323566e-03 1 ## Pex3 1.085048e-07 0.4162320 0.518 0.317 4.329886e-03 1 ## Cspg5 1.095430e-07 0.5925473 0.261 0.122 4.371312e-03 1 ## Eif3g1 1.108536e-07 0.3294544 0.788 0.484 4.423614e-03 1 ## Vapa1 1.115604e-07 0.2192248 0.959 0.755 4.451819e-03 1 ## Cd274 1.116445e-07 1.1482490 0.098 0.027 4.455174e-03 1 ## U2af1l41 1.125038e-07 0.2132194 0.563 0.336 4.489464e-03 1 ## Gm32385 1.126866e-07 2.2539295 0.049 0.007 4.496759e-03 1 ## Ptpdc1 1.131358e-07 1.1890148 0.082 0.020 4.514685e-03 1 ## Zcrb1 1.134222e-07 0.2492005 0.906 0.659 4.526112e-03 1 ## Gkap1 1.152709e-07 0.4853712 0.327 0.171 4.599887e-03 1 ## Gtf2i1 1.153740e-07 0.1998419 0.812 0.539 4.603999e-03 1 ## Arfgap1 1.153799e-07 0.2823552 0.510 0.303 4.604235e-03 1 ## Onecut21 1.167229e-07 0.1335993 0.996 0.789 4.657829e-03 1 ## Selenoh 1.172571e-07 0.2010797 0.290 0.140 4.679145e-03 1 ## Bag5 1.175331e-07 0.7152723 0.339 0.182 4.690159e-03 1 ## Sumo21 1.189504e-07 0.2922139 0.963 0.738 4.746715e-03 1 ## Sgtb 1.209092e-07 0.5917857 0.229 0.102 4.824881e-03 1 ## Mrps331 1.215989e-07 0.2762707 0.943 0.676 4.852405e-03 1 ## Pigg 1.216538e-07 0.5785634 0.273 0.132 4.854596e-03 1 ## Tsen34 1.219267e-07 0.2935513 0.714 0.461 4.865485e-03 1 ## E130311K13Rik 1.229622e-07 0.6419651 0.273 0.129 4.906808e-03 1 ## 2410002F23Rik 1.239194e-07 0.2021183 0.359 0.191 4.945002e-03 1 ## Dusp22 1.248136e-07 0.8029997 0.163 0.062 4.980685e-03 1 ## Polr3d 1.258234e-07 0.4805362 0.408 0.236 5.020982e-03 1 ## Rpl36al 1.258699e-07 0.1751923 1.000 0.835 5.022840e-03 1 ## Gm10033 1.286754e-07 0.4605837 0.273 0.132 5.134790e-03 1 ## Smad2 1.289538e-07 0.3721828 0.473 0.274 5.145902e-03 1 ## Gm35534 1.292996e-07 2.6543201 0.037 0.003 5.159701e-03 1 ## Gm2137 1.293739e-07 1.3331730 0.110 0.034 5.162666e-03 1 ## Fggy 1.294724e-07 0.8717578 0.208 0.090 5.166597e-03 1 ## Sh3bgrl 1.304902e-07 0.1808591 0.918 0.660 5.207210e-03 1 ## Rp9 1.306210e-07 0.1924211 0.808 0.553 5.212431e-03 1 ## Pdhb 1.310684e-07 0.4303202 0.641 0.410 5.230284e-03 1 ## Elp3 1.318193e-07 0.5183052 0.371 0.201 5.260248e-03 1 ## Zfp617 1.318763e-07 0.5125010 0.376 0.209 5.262525e-03 1 ## Isca2 1.327231e-07 0.5649439 0.494 0.299 5.296316e-03 1 ## Gm320171 1.336654e-07 0.7033466 0.131 0.043 5.333917e-03 1 ## Dpm1 1.338962e-07 0.6499070 0.363 0.198 5.343129e-03 1 ## Mrpl13 1.352426e-07 0.1602819 0.527 0.298 5.396855e-03 1 ## Tm9sf2 1.360471e-07 0.2036461 0.943 0.687 5.428961e-03 1 ## Zc3h7b1 1.388727e-07 0.4568836 0.453 0.256 5.541716e-03 1 ## Gsk3b1 1.404770e-07 0.2252671 0.914 0.702 5.605736e-03 1 ## Gm36318 1.455028e-07 0.3464473 0.392 0.220 5.806288e-03 1 ## Gm40543 1.481304e-07 1.3177536 0.090 0.024 5.911144e-03 1 ## Atp5pd1 1.496051e-07 0.1525450 1.000 0.858 5.969991e-03 1 ## Nudt141 1.501874e-07 0.3624829 0.735 0.500 5.993227e-03 1 ## Kdm1b 1.521492e-07 0.5945836 0.278 0.138 6.071516e-03 1 ## Cibar11 1.523578e-07 0.5825818 0.392 0.221 6.079837e-03 1 ## Lyset1 1.540058e-07 0.5944001 0.367 0.201 6.145602e-03 1 ## Ankrd24 1.543260e-07 0.4127701 0.298 0.147 6.158379e-03 1 ## Psma11 1.550146e-07 0.2362417 0.845 0.592 6.185857e-03 1 ## Brk11 1.554034e-07 0.3081479 0.918 0.675 6.201373e-03 1 ## Ssr1 1.562373e-07 0.2510563 0.898 0.641 6.234651e-03 1 ## Mcf2l1 1.570848e-07 0.2124049 0.853 0.612 6.268471e-03 1 ## Xpa 1.579800e-07 0.3193337 0.555 0.336 6.304191e-03 1 ## Actl6b 1.590759e-07 0.5599025 0.282 0.136 6.347924e-03 1 ## Med14 1.593253e-07 0.2806805 0.465 0.275 6.357876e-03 1 ## Ywhae1 1.594825e-07 0.2795432 1.000 0.826 6.364151e-03 1 ## Tial1 1.600885e-07 0.2633475 0.837 0.593 6.388332e-03 1 ## Thoc2 1.603585e-07 0.2087731 0.935 0.719 6.399107e-03 1 ## Nol12 1.613672e-07 0.3188655 0.498 0.287 6.439359e-03 1 ## Pmvk 1.618884e-07 0.4434181 0.510 0.308 6.460156e-03 1 ## Rfx7 1.619987e-07 0.2654949 0.522 0.305 6.464559e-03 1 ## Ndufa111 1.645981e-07 0.1823109 0.963 0.746 6.568285e-03 1 ## Asah11 1.654822e-07 0.2149880 0.755 0.500 6.603567e-03 1 ## Prkx1 1.676690e-07 0.3115882 0.649 0.398 6.690833e-03 1 ## Rab141 1.678729e-07 0.2298206 0.980 0.791 6.698968e-03 1 ## Ubap1l 1.692541e-07 0.4707440 0.110 0.034 6.754085e-03 1 ## Gm45916 1.699747e-07 0.8372686 0.196 0.083 6.782841e-03 1 ## Chkb 1.727314e-07 0.3359577 0.376 0.201 6.892848e-03 1 ## Slc17a9 1.730115e-07 0.7963105 0.082 0.020 6.904026e-03 1 ## Fbxw9 1.731845e-07 0.3872254 0.400 0.220 6.910927e-03 1 ## Abhd14b 1.761948e-07 0.3035604 0.653 0.433 7.031054e-03 1 ## Zfp52 1.769967e-07 0.5346473 0.196 0.080 7.063055e-03 1 ## Polr2m1 1.781544e-07 0.2595716 0.865 0.601 7.109253e-03 1 ## Gm40150 1.791684e-07 1.1600386 0.094 0.026 7.149713e-03 1 ## Slc39a11 1.801478e-07 0.3504158 0.453 0.259 7.188799e-03 1 ## Fads11 1.805384e-07 0.5431247 0.416 0.247 7.204386e-03 1 ## Tlcd4 1.816835e-07 0.3040940 0.404 0.229 7.250080e-03 1 ## Rab321 1.827634e-07 0.6317657 0.249 0.119 7.293175e-03 1 ## Fbxo17 1.843434e-07 1.1901269 0.082 0.020 7.356222e-03 1 ## Chst11 1.851037e-07 0.9559427 0.163 0.064 7.386563e-03 1 ## Gm32849 1.899602e-07 2.6122532 0.061 0.012 7.580361e-03 1 ## Cyren 1.901758e-07 0.6064053 0.220 0.101 7.588964e-03 1 ## Bcl7a 1.910753e-07 0.3052234 0.461 0.265 7.624860e-03 1 ## Gmds 1.919042e-07 0.2699577 0.857 0.571 7.657936e-03 1 ## Ptprg1 1.924157e-07 0.2257722 0.641 0.412 7.678348e-03 1 ## Rbx11 1.974171e-07 0.2198395 0.971 0.767 7.877930e-03 1 ## Tlcd3a1 1.976590e-07 0.4191547 0.302 0.155 7.887582e-03 1 ## Fam83d1 1.991492e-07 0.5115387 0.249 0.119 7.947048e-03 1 ## Selenon 2.013498e-07 0.5077176 0.302 0.151 8.034863e-03 1 ## Nudt19 2.042363e-07 0.3001432 0.490 0.273 8.150048e-03 1 ## Dnajc31 2.043210e-07 0.2670976 0.922 0.683 8.153431e-03 1 ## Ypel1 2.060233e-07 1.6028121 0.073 0.017 8.221359e-03 1 ## Tmem216 2.065365e-07 0.8677500 0.241 0.115 8.241840e-03 1 ## Gpr75 2.094447e-07 1.0752434 0.106 0.032 8.357889e-03 1 ## Cd79b 2.127485e-07 2.6547577 0.057 0.010 8.489729e-03 1 ## Dnajc30 2.150657e-07 0.4441573 0.355 0.189 8.582195e-03 1 ## Gopc 2.153987e-07 0.3406057 0.510 0.295 8.595483e-03 1 ## G630025P09Rik 2.170771e-07 0.8791932 0.176 0.071 8.662463e-03 1 ## Chchd7 2.204100e-07 0.2981760 0.641 0.405 8.795461e-03 1 ## Rabggtb 2.208360e-07 0.1819325 0.518 0.321 8.812462e-03 1 ## Pcdhb4 2.223428e-07 2.4316556 0.057 0.010 8.872587e-03 1 ## Itpripl2 2.224837e-07 0.5975311 0.253 0.124 8.878212e-03 1 ## Gm46617 2.242836e-07 0.3632158 0.290 0.143 8.950037e-03 1 ## Cmc4 2.272545e-07 0.8121433 0.261 0.131 9.068592e-03 1 ## Palm 2.279915e-07 1.3704612 0.114 0.036 9.098003e-03 1 ## Zfp40 2.287770e-07 0.2949436 0.278 0.137 9.129346e-03 1 ## Acacb 2.328323e-07 1.1751699 0.078 0.019 9.291174e-03 1 ## Golga71 2.337557e-07 0.5410206 0.494 0.291 9.328021e-03 1 ## Slc25a12 2.340035e-07 1.0256475 0.159 0.063 9.337909e-03 1 ## Mdc1 2.370435e-07 0.3506758 0.314 0.162 9.459221e-03 1 ## Pten1 2.370836e-07 0.2451958 0.971 0.742 9.460820e-03 1 ## Gdap21 2.394630e-07 0.6221854 0.310 0.157 9.555770e-03 1 ## Abhd17a 2.410446e-07 0.2191926 0.841 0.569 9.618883e-03 1 ## BC051226 2.410512e-07 0.9304004 0.212 0.097 9.619147e-03 1 ## Krt101 2.427285e-07 0.1808786 0.457 0.272 9.686081e-03 1 ## Hook1 2.433294e-07 0.2463258 0.776 0.528 9.710059e-03 1 ## Fbxl20 2.439812e-07 0.4001693 0.404 0.228 9.736069e-03 1 ## Cdh4 2.440318e-07 0.9752102 0.131 0.045 9.738091e-03 1 ## Mfsd4b1 2.461860e-07 0.6554923 0.216 0.094 9.824050e-03 1 ## Naa35 2.470841e-07 0.2598901 0.690 0.435 9.859893e-03 1 ## Smim20 2.542555e-07 0.3764695 0.608 0.370 1.014607e-02 1 ## Abtb11 2.555895e-07 0.2232729 0.457 0.254 1.019930e-02 1 ## Tsc22d3 2.564917e-07 0.3622071 0.257 0.128 1.023530e-02 1 ## Id31 2.586914e-07 0.3313032 0.980 0.857 1.032308e-02 1 ## 0610040B10Rik 2.590708e-07 0.8891793 0.204 0.089 1.033822e-02 1 ## 4930503L19Rik 2.604518e-07 1.0255032 0.139 0.050 1.039333e-02 1 ## Wbp41 2.608450e-07 0.2546845 0.812 0.558 1.040902e-02 1 ## Gys1 2.676298e-07 0.6992287 0.167 0.067 1.067977e-02 1 ## Pelp1 2.705741e-07 0.4351764 0.343 0.182 1.079726e-02 1 ## Herc3 2.722566e-07 0.5938880 0.351 0.196 1.086440e-02 1 ## Gm15816 2.723019e-07 2.1083636 0.053 0.009 1.086621e-02 1 ## Zfp950 2.748692e-07 0.1615172 0.796 0.561 1.096866e-02 1 ## Nfu11 2.761642e-07 0.2568776 0.555 0.342 1.102033e-02 1 ## Ift74 2.790519e-07 0.4992005 0.310 0.162 1.113557e-02 1 ## Fcor 2.805347e-07 1.5075348 0.094 0.027 1.119474e-02 1 ## Suz12 2.822097e-07 0.2383968 0.763 0.526 1.126158e-02 1 ## H2bc271 2.826234e-07 0.6443961 0.278 0.138 1.127809e-02 1 ## Msantd41 2.875580e-07 0.2632891 0.657 0.427 1.147500e-02 1 ## Suv39h1 2.915094e-07 0.5515044 0.196 0.087 1.163268e-02 1 ## Dph3 2.931534e-07 0.2440863 0.776 0.521 1.169829e-02 1 ## Anxa4 2.957481e-07 0.1501095 0.992 0.897 1.180183e-02 1 ## Kifbp 2.964240e-07 0.5821274 0.482 0.294 1.182880e-02 1 ## Ndufb71 3.029139e-07 0.1344340 0.992 0.806 1.208778e-02 1 ## AI480526 3.063173e-07 0.1361603 0.539 0.313 1.222359e-02 1 ## Tmem50a1 3.074002e-07 0.3121007 0.963 0.727 1.226681e-02 1 ## Cyp2j8 3.106016e-07 2.6802299 0.033 0.003 1.239456e-02 1 ## Dguok 3.125285e-07 0.4043280 0.518 0.317 1.247145e-02 1 ## Gm36220 3.129668e-07 0.4349837 0.318 0.166 1.248894e-02 1 ## Ifih11 3.138037e-07 0.1793064 0.600 0.374 1.252234e-02 1 ## Polr2e1 3.151920e-07 0.3332443 0.727 0.491 1.257774e-02 1 ## Hibch 3.287495e-07 0.2280296 0.306 0.160 1.311875e-02 1 ## Spata24 3.332130e-07 1.4052962 0.139 0.053 1.329686e-02 1 ## Gm35611 3.339302e-07 2.0479179 0.065 0.014 1.332548e-02 1 ## Ehd4 3.398110e-07 0.4666893 0.282 0.145 1.356016e-02 1 ## Nt5c1 3.426387e-07 0.2375984 0.943 0.692 1.367300e-02 1 ## Arhgap29 3.435812e-07 1.0352242 0.167 0.070 1.371061e-02 1 ## Cnn31 3.445970e-07 0.2180713 0.837 0.570 1.375114e-02 1 ## Znrf3 3.479218e-07 1.0292422 0.180 0.078 1.388382e-02 1 ## Ppp5c 3.497539e-07 0.4316307 0.527 0.322 1.395693e-02 1 ## A930001C03Rik 3.524870e-07 2.1315974 0.065 0.014 1.406599e-02 1 ## Lgals1 3.525903e-07 2.6519574 0.061 0.013 1.407012e-02 1 ## Lipa 3.543339e-07 0.1677404 0.388 0.205 1.413969e-02 1 ## Aip1 3.546216e-07 0.3812946 0.563 0.347 1.415117e-02 1 ## Pdk1 3.580126e-07 0.6093107 0.302 0.162 1.428649e-02 1 ## Gm7932 3.612480e-07 1.1099037 0.098 0.029 1.441560e-02 1 ## Acer1 3.664452e-07 0.4720735 0.384 0.212 1.462299e-02 1 ## Pnkd1 3.670698e-07 0.2193473 0.800 0.519 1.464792e-02 1 ## A430005L14Rik 3.676497e-07 0.4819376 0.408 0.229 1.467106e-02 1 ## Uba21 3.747267e-07 0.2476352 0.596 0.363 1.495347e-02 1 ## Thap111 3.769617e-07 0.4779507 0.384 0.208 1.504266e-02 1 ## Manba 3.770422e-07 1.3350840 0.159 0.065 1.504587e-02 1 ## D730005E14Rik 3.879281e-07 0.2176432 0.127 0.045 1.548027e-02 1 ## Cdk5rap3 3.925763e-07 0.2180512 0.702 0.451 1.566576e-02 1 ## AW1120101 3.935872e-07 0.3965309 0.637 0.423 1.570610e-02 1 ## Lsm5 3.971135e-07 0.1543804 0.465 0.261 1.584682e-02 1 ## Cfap52 3.985454e-07 1.8535614 0.065 0.014 1.590396e-02 1 ## Gatd1 3.996484e-07 0.3873754 0.461 0.272 1.594797e-02 1 ## C630043F03Rik 4.010553e-07 0.7309072 0.208 0.094 1.600411e-02 1 ## Dnm1l 4.010933e-07 0.1886032 0.812 0.565 1.600563e-02 1 ## Uggt2 4.026838e-07 1.5224220 0.102 0.031 1.606910e-02 1 ## Cnksr3 4.043968e-07 0.3615932 0.339 0.178 1.613746e-02 1 ## Bcl11b 4.068006e-07 0.2260723 0.498 0.289 1.623338e-02 1 ## Grb7 4.079256e-07 0.2706310 0.690 0.432 1.627827e-02 1 ## Elk3 4.103997e-07 0.9776703 0.143 0.054 1.637700e-02 1 ## Fkbp11 4.117273e-07 0.8237073 0.196 0.088 1.642998e-02 1 ## Shroom1 4.130812e-07 1.2787594 0.118 0.040 1.648400e-02 1 ## Ttc32 4.155368e-07 0.5313394 0.400 0.229 1.658200e-02 1 ## Zfp593 4.194036e-07 0.8106475 0.261 0.130 1.673630e-02 1 ## Hcn3 4.209214e-07 0.9658957 0.122 0.041 1.679687e-02 1 ## 2210406O10Rik 4.305565e-07 2.0721460 0.061 0.013 1.718136e-02 1 ## Sft2d1 4.464428e-07 0.1422393 0.473 0.273 1.781530e-02 1 ## Chid11 4.489230e-07 0.4095413 0.498 0.294 1.791427e-02 1 ## Mrpl35 4.511699e-07 0.3476210 0.535 0.304 1.800394e-02 1 ## Tmem250 4.512961e-07 0.5175884 0.433 0.253 1.800897e-02 1 ## Wwp1 4.608789e-07 0.3190939 0.469 0.282 1.839137e-02 1 ## Armcx3 4.628030e-07 0.4779198 0.408 0.238 1.846815e-02 1 ## Fgfbp3 4.740845e-07 0.7894371 0.261 0.129 1.891834e-02 1 ## Tnrc6b 4.756107e-07 0.1371354 0.922 0.671 1.897924e-02 1 ## Commd3 4.766772e-07 0.2354331 0.604 0.363 1.902180e-02 1 ## Calhm21 4.775063e-07 0.9706798 0.131 0.045 1.905489e-02 1 ## Zdhhc8 4.784014e-07 0.2000658 0.343 0.181 1.909061e-02 1 ## Polr2f1 4.896518e-07 0.1547875 0.910 0.628 1.953955e-02 1 ## Fam120c 4.950258e-07 0.5518740 0.363 0.199 1.975400e-02 1 ## Cops7a 4.951180e-07 0.4143122 0.608 0.386 1.975769e-02 1 ## Mfsd9 4.959272e-07 0.5057515 0.245 0.115 1.978997e-02 1 ## Hilpda 4.982856e-07 1.4230808 0.163 0.068 1.988409e-02 1 ## Bccip 5.013628e-07 0.1749472 0.673 0.433 2.000688e-02 1 ## Tmem141 5.066003e-07 0.4791956 0.335 0.178 2.021588e-02 1 ## Gm57587 5.080144e-07 1.2323689 0.122 0.042 2.027231e-02 1 ## Zfp637 5.156923e-07 0.3679333 0.396 0.229 2.057870e-02 1 ## Zfp551 5.161517e-07 0.9298080 0.155 0.061 2.059703e-02 1 ## Zmynd19 5.212462e-07 0.6226317 0.196 0.087 2.080033e-02 1 ## Hipk3 5.214525e-07 0.3269614 0.633 0.410 2.080856e-02 1 ## Krcc11 5.252197e-07 0.2245381 0.869 0.629 2.095889e-02 1 ## A830021M18Rik 5.407742e-07 3.4643187 0.037 0.004 2.157960e-02 1 ## Septin8 5.456873e-07 0.7523586 0.216 0.101 2.177565e-02 1 ## Wrn 5.463816e-07 0.4916983 0.339 0.185 2.180336e-02 1 ## Smarcb11 5.499203e-07 0.3122273 0.649 0.414 2.194457e-02 1 ## Tsen15 5.536468e-07 0.8956823 0.233 0.112 2.209328e-02 1 ## Vps4b1 5.612774e-07 0.1731074 0.837 0.564 2.239778e-02 1 ## Aldh7a11 5.673177e-07 0.5196079 0.551 0.351 2.263881e-02 1 ## Snhg10 5.706812e-07 0.6190507 0.135 0.050 2.277303e-02 1 ## Gm40656 5.710548e-07 0.3975301 0.229 0.105 2.278794e-02 1 ## Tm9sf41 5.720855e-07 0.2755514 0.702 0.473 2.282907e-02 1 ## Slc25a17 5.724832e-07 0.3060531 0.416 0.249 2.284494e-02 1 ## Arhgef281 5.740608e-07 0.2257629 0.641 0.422 2.290789e-02 1 ## Pdcd101 5.765496e-07 0.2118819 0.841 0.577 2.300721e-02 1 ## Unc13a1 5.861395e-07 0.2033582 0.498 0.290 2.338990e-02 1 ## Katnbl1 5.875365e-07 0.4015094 0.331 0.177 2.344565e-02 1 ## Fkbp14 5.898808e-07 1.3365173 0.102 0.032 2.353919e-02 1 ## Rwdd1 5.901142e-07 0.2158005 0.804 0.540 2.354851e-02 1 ## Dcakd 5.918558e-07 0.6055655 0.441 0.268 2.361801e-02 1 ## Postn 5.925710e-07 2.1494714 0.057 0.011 2.364655e-02 1 ## Brdt 5.965899e-07 0.3816873 0.184 0.079 2.380692e-02 1 ## Nrbp2 5.972650e-07 0.7314819 0.216 0.102 2.383386e-02 1 ## Pdrg1 6.008645e-07 0.4843926 0.433 0.253 2.397750e-02 1 ## Rps6ka2 6.008776e-07 0.2425101 0.290 0.143 2.397802e-02 1 ## Mppe1 6.083000e-07 0.5868278 0.327 0.182 2.427421e-02 1 ## Ifngr11 6.105658e-07 0.2468090 0.682 0.445 2.436463e-02 1 ## Gatad2b 6.133854e-07 0.2643202 0.804 0.572 2.447715e-02 1 ## Tmem135 6.147322e-07 0.1503297 0.608 0.377 2.453089e-02 1 ## Dnah7b 6.168909e-07 2.7449118 0.037 0.004 2.461703e-02 1 ## Pds5b 6.279558e-07 0.2492824 0.441 0.264 2.505858e-02 1 ## Cep170b1 6.293403e-07 0.2616750 0.690 0.457 2.511382e-02 1 ## Prr15l 6.396505e-07 0.2271487 0.971 0.736 2.552525e-02 1 ## Prpf6 6.473937e-07 0.1470900 0.527 0.323 2.583424e-02 1 ## Stat5a 6.502800e-07 0.5186871 0.139 0.052 2.594942e-02 1 ## Pam1 6.536421e-07 0.2859709 0.996 0.800 2.608359e-02 1 ## Scand11 6.566344e-07 0.2406436 0.980 0.776 2.620300e-02 1 ## Gm57735 6.566996e-07 0.6755781 0.212 0.098 2.620560e-02 1 ## Slc16a6 6.624785e-07 0.3430227 0.327 0.176 2.643620e-02 1 ## 5430405H02Rik 6.625231e-07 0.3362791 0.290 0.149 2.643798e-02 1 ## Gemin71 6.628318e-07 0.3775837 0.559 0.344 2.645030e-02 1 ## Thoc7 6.631177e-07 0.1209138 0.902 0.650 2.646171e-02 1 ## Iah1 6.645354e-07 0.2863673 0.502 0.310 2.651829e-02 1 ## Srgap2 6.655604e-07 0.3126350 0.286 0.151 2.655919e-02 1 ## Tacc1 6.668014e-07 0.2447974 0.420 0.235 2.660871e-02 1 ## Pde6d1 6.743628e-07 0.3321682 0.486 0.296 2.691045e-02 1 ## Septin71 6.777936e-07 0.2238100 0.922 0.648 2.704735e-02 1 ## Gm30789 6.782800e-07 1.2335171 0.102 0.032 2.706676e-02 1 ## Sema4d 6.785158e-07 0.8834353 0.180 0.077 2.707617e-02 1 ## Tsn1 6.805245e-07 0.2445466 0.861 0.605 2.715633e-02 1 ## Nudt11 6.809483e-07 0.9861689 0.159 0.065 2.717324e-02 1 ## Celf31 6.825378e-07 0.1829850 0.984 0.778 2.723667e-02 1 ## Gm51920 6.867401e-07 1.3135381 0.106 0.034 2.740437e-02 1 ## Slc41a1 7.133268e-07 0.7701746 0.184 0.081 2.846530e-02 1 ## Eea1 7.154609e-07 0.1097491 0.865 0.632 2.855047e-02 1 ## Elp21 7.161623e-07 0.2683298 0.392 0.224 2.857846e-02 1 ## 1700048O20Rik 7.165327e-07 2.6858907 0.041 0.006 2.859324e-02 1 ## Ift52 7.181469e-07 0.4221583 0.416 0.250 2.865765e-02 1 ## Sod2 7.303431e-07 0.2105982 0.853 0.593 2.914434e-02 1 ## Usp53 7.329835e-07 0.3268666 0.437 0.256 2.924971e-02 1 ## Sh3glb11 7.411908e-07 0.1758845 0.984 0.803 2.957722e-02 1 ## Gm53687 7.467092e-07 1.4109290 0.053 0.010 2.979743e-02 1 ## Cep112it 7.544403e-07 2.6995670 0.049 0.008 3.010594e-02 1 ## Frs3 7.672467e-07 0.6904603 0.224 0.106 3.061698e-02 1 ## Ggnbp2os 7.692586e-07 0.3958676 0.412 0.244 3.069726e-02 1 ## Eml1 7.712425e-07 0.3375445 0.392 0.218 3.077643e-02 1 ## Pink1 7.864864e-07 0.2318234 0.678 0.427 3.138474e-02 1 ## 4930517O19Rik 7.930132e-07 2.4815536 0.049 0.008 3.164519e-02 1 ## Cap11 8.000436e-07 0.1387373 0.743 0.492 3.192574e-02 1 ## Ciao2a1 8.029899e-07 0.1619375 0.788 0.507 3.204331e-02 1 ## Tada3 8.083402e-07 0.4241310 0.396 0.232 3.225681e-02 1 ## Tmem60 8.139101e-07 0.4123022 0.441 0.254 3.247908e-02 1 ## Skic3 8.147567e-07 0.3505257 0.327 0.179 3.251287e-02 1 ## Paupar 8.156193e-07 0.1292038 0.367 0.198 3.254729e-02 1 ## Siah1a 8.184823e-07 0.2646002 0.510 0.311 3.266154e-02 1 ## Tmx3 8.203822e-07 0.2132478 0.604 0.385 3.273735e-02 1 ## Galnt11 8.278997e-07 0.2969315 0.457 0.259 3.303734e-02 1 ## Brix1 8.346391e-07 0.2720065 0.449 0.266 3.330627e-02 1 ## Cops5 8.458979e-07 0.3227447 0.714 0.487 3.375556e-02 1 ## Rab431 8.566254e-07 0.2914184 0.604 0.387 3.418364e-02 1 ## Angel1 8.569259e-07 0.8194770 0.155 0.061 3.419563e-02 1 ## Sdc1 8.641001e-07 0.3324053 0.310 0.164 3.448191e-02 1 ## Gm11963 8.656408e-07 0.5763066 0.208 0.093 3.454340e-02 1 ## Ankrd2 8.659312e-07 2.3092275 0.045 0.007 3.455499e-02 1 ## Cox6c1 8.674544e-07 0.1175436 1.000 0.871 3.461577e-02 1 ## Map3k12 8.784051e-07 0.3690312 0.237 0.111 3.505276e-02 1 ## Terf1 8.797113e-07 0.4588865 0.363 0.200 3.510488e-02 1 ## 1810019D21Rik 8.809955e-07 0.5489444 0.286 0.150 3.515613e-02 1 ## Mkrn11 8.854252e-07 0.2125421 0.922 0.642 3.533289e-02 1 ## Bola11 8.914790e-07 0.3287899 0.531 0.312 3.557447e-02 1 ## Snx24 8.970054e-07 0.4451471 0.420 0.249 3.579500e-02 1 ## Golga3 9.073405e-07 0.2160158 0.510 0.307 3.620742e-02 1 ## Nup88 9.132703e-07 0.2748030 0.527 0.331 3.644405e-02 1 ## Ripor1 9.135075e-07 0.6013074 0.282 0.150 3.645352e-02 1 ## Golt1a 9.176999e-07 0.1182772 0.735 0.486 3.662082e-02 1 ## Babam21 9.212450e-07 0.4777365 0.408 0.242 3.676228e-02 1 ## Trafd1 9.215162e-07 0.2330666 0.629 0.403 3.677311e-02 1 ## Notch1 9.287797e-07 1.0253954 0.171 0.075 3.706295e-02 1 ## Dancr 9.292212e-07 0.4753176 0.212 0.098 3.708057e-02 1 ## Ddx50 9.299844e-07 0.1395410 0.739 0.506 3.711103e-02 1 ## Slain1 9.405403e-07 1.0589715 0.180 0.080 3.753226e-02 1 ## Emc6 9.499277e-07 0.2402421 0.673 0.435 3.790687e-02 1 ## Cct21 9.537818e-07 0.1806985 0.922 0.668 3.806066e-02 1 ## 2610029K11Rik 9.614485e-07 1.9626461 0.098 0.031 3.836660e-02 1 ## Comt 9.628685e-07 0.2383252 0.837 0.570 3.842327e-02 1 ## Ube2j1 9.733178e-07 0.2296740 0.510 0.320 3.884025e-02 1 ## Faf21 9.810887e-07 0.3276723 0.722 0.475 3.915034e-02 1 ## Pafah1b21 9.988633e-07 0.3219660 0.722 0.481 3.985964e-02 1 ## Picalm 9.990130e-07 0.1710518 0.808 0.565 3.986561e-02 1 ## Tmem260 1.002428e-06 0.3447472 0.314 0.168 4.000190e-02 1 ## Hnrnpr 1.002754e-06 0.2434710 0.837 0.592 4.001492e-02 1 ## Zfp3261 1.033025e-06 0.2083859 0.755 0.535 4.122285e-02 1 ## Rsl1d11 1.070267e-06 0.1271685 0.604 0.363 4.270902e-02 1 ## Gm49870 1.079233e-06 1.1477622 0.102 0.033 4.306679e-02 1 ## Zfp949 1.079687e-06 0.3879662 0.367 0.206 4.308490e-02 1 ## Cd59b1 1.081393e-06 0.4519525 0.290 0.147 4.315300e-02 1 ## Fh1 1.083675e-06 0.1446101 0.592 0.352 4.324405e-02 1 ## Ddx11 1.087871e-06 0.1704561 0.878 0.618 4.341150e-02 1 ## Gm53710 1.098624e-06 1.3976617 0.110 0.037 4.384060e-02 1 ## Tmem87b 1.103110e-06 0.2196725 0.694 0.454 4.401961e-02 1 ## Sertad3 1.125430e-06 0.5835303 0.241 0.119 4.491027e-02 1 ## Pigf 1.125925e-06 0.4955174 0.420 0.249 4.493004e-02 1 ## Gga2 1.129365e-06 0.7171143 0.212 0.101 4.506730e-02 1 ## Kctd201 1.129495e-06 0.1858365 0.543 0.336 4.507250e-02 1 ## Mdp1 1.130825e-06 0.3447564 0.608 0.389 4.512555e-02 1 ## Ildr1 1.144175e-06 0.1551596 0.563 0.337 4.565831e-02 1 ## Tomm70a1 1.145027e-06 0.2649529 0.743 0.492 4.569230e-02 1 ## Nsfl1c1 1.170861e-06 0.2383947 0.702 0.451 4.672319e-02 1 ## Adgb 1.174840e-06 0.1194262 0.265 0.130 4.688199e-02 1 ## Dtx3 1.177999e-06 0.1885890 0.380 0.203 4.700804e-02 1 ## Cfap68 1.192157e-06 0.2028653 0.531 0.326 4.757301e-02 1 ## 4930579K19Rik 1.210807e-06 0.6444434 0.241 0.119 4.831727e-02 1 ## Usp48 1.214951e-06 0.1058302 0.739 0.504 4.848263e-02 1 ## Mrps211 1.217346e-06 0.1975405 0.918 0.627 4.857820e-02 1 ## Gm17202 1.217399e-06 0.4651274 0.147 0.058 4.858031e-02 1 ## Cfl2 1.229168e-06 0.1814644 0.624 0.403 4.904994e-02 1 ## Enc1 1.232548e-06 0.4471785 0.343 0.191 4.918484e-02 1 ## Mrps26 1.233857e-06 0.2856770 0.629 0.396 4.923708e-02 1 ## Gm17576 1.244102e-06 3.7643982 0.024 0.001 4.964588e-02 1 ## Gm36620 1.259197e-06 3.5944510 0.024 0.001 5.024824e-02 1 ## Usf1 1.266344e-06 0.3615170 0.461 0.283 5.053345e-02 1 ## Sae1 1.275676e-06 0.1088166 0.494 0.294 5.090585e-02 1 ## Wdr77 1.282905e-06 0.3813139 0.355 0.197 5.119431e-02 1 ## Mrpl14 1.286674e-06 0.1155920 0.841 0.556 5.134472e-02 1 ## Rtn41 1.289278e-06 0.1376379 0.939 0.718 5.144864e-02 1 ## Add3 1.293383e-06 0.1407290 0.792 0.505 5.161244e-02 1 ## Mov10 1.294922e-06 0.1701312 0.596 0.370 5.167385e-02 1 ## Vegfd 1.297709e-06 3.2104039 0.024 0.001 5.178508e-02 1 ## Plod3 1.298443e-06 0.6157896 0.310 0.171 5.181438e-02 1 ## Adgrb3 1.305546e-06 3.2851816 0.024 0.001 5.209782e-02 1 ## Oprl1 1.305546e-06 2.4565924 0.024 0.001 5.209782e-02 1 ## Gm46119 1.313429e-06 2.2661088 0.024 0.001 5.241239e-02 1 ## Crhbp 1.313429e-06 2.8385124 0.024 0.001 5.241239e-02 1 ## Gm53575 1.317862e-06 3.7491288 0.016 0.000 5.258929e-02 1 ## Gm53488 1.317862e-06 3.7657124 0.016 0.000 5.258929e-02 1 ## Usp13 1.317862e-06 3.9568303 0.016 0.000 5.258929e-02 1 ## Gm16499 1.317862e-06 3.7750281 0.016 0.000 5.258929e-02 1 ## Ccdc179 1.317862e-06 4.1930921 0.016 0.000 5.258929e-02 1 ## 9130015A21Rik 1.317862e-06 3.7754471 0.016 0.000 5.258929e-02 1 ## Gm34766 1.317862e-06 4.0286387 0.016 0.000 5.258929e-02 1 ## Adamts10 1.317862e-06 3.7876176 0.016 0.000 5.258929e-02 1 ## Cyp2c37 1.317862e-06 4.0755825 0.016 0.000 5.258929e-02 1 ## Tlr8 1.317862e-06 3.7343190 0.016 0.000 5.258929e-02 1 ## Slc25a47 1.318378e-06 1.0055864 0.135 0.051 5.260986e-02 1 ## Uevld 1.321096e-06 0.7026937 0.249 0.125 5.271834e-02 1 ## B9d2 1.321860e-06 0.5645950 0.278 0.142 5.274883e-02 1 ## Stx1a1 1.330630e-06 0.3053102 0.576 0.363 5.309878e-02 1 ## Extl3 1.353995e-06 0.4220720 0.347 0.196 5.403116e-02 1 ## Bbln1 1.365029e-06 0.2528196 0.747 0.503 5.447149e-02 1 ## Psmb41 1.365768e-06 0.1839893 0.976 0.749 5.450097e-02 1 ## 2810408B13Rik 1.369015e-06 0.8952476 0.135 0.052 5.463053e-02 1 ## Spryd3 1.405819e-06 0.2023523 0.371 0.213 5.609919e-02 1 ## Ppp2r5a 1.407096e-06 0.2161858 0.698 0.463 5.615018e-02 1 ## BC005624 1.434026e-06 0.2518104 0.563 0.363 5.722481e-02 1 ## Enpp1 1.439679e-06 0.1517538 0.163 0.068 5.745039e-02 1 ## Bbip1 1.450192e-06 0.2604045 0.682 0.453 5.786991e-02 1 ## Gm39874 1.453337e-06 1.7190292 0.094 0.029 5.799540e-02 1 ## Arf5 1.472225e-06 0.2250099 0.996 0.834 5.874912e-02 1 ## Trim34a 1.476351e-06 0.7271752 0.151 0.061 5.891379e-02 1 ## Trmt12 1.479986e-06 0.6575910 0.208 0.097 5.905884e-02 1 ## Gm19531 1.488668e-06 0.6680127 0.143 0.055 5.940529e-02 1 ## Mkrn2 1.490513e-06 0.4080928 0.286 0.150 5.947894e-02 1 ## Camk2g 1.497947e-06 0.7956967 0.237 0.118 5.977556e-02 1 ## Osgep1 1.512707e-06 0.3051996 0.608 0.389 6.036456e-02 1 ## Acp21 1.513439e-06 0.1908013 0.580 0.359 6.039376e-02 1 ## Actr8 1.535127e-06 0.4507977 0.327 0.182 6.125923e-02 1 ## Gm35685 1.540090e-06 0.2397363 0.400 0.218 6.145730e-02 1 ## Rnf411 1.545079e-06 0.1934574 0.494 0.296 6.165638e-02 1 ## Pals2 1.547685e-06 0.1773287 0.527 0.316 6.176038e-02 1 ## Phtf2 1.548093e-06 0.6566005 0.143 0.056 6.177667e-02 1 ## Cdkn2aipnl 1.555328e-06 0.4326152 0.351 0.199 6.206535e-02 1 ## Uqcrb1 1.557552e-06 0.1227122 0.988 0.774 6.215413e-02 1 ## Ccdc1481 1.559565e-06 0.5662491 0.192 0.086 6.223444e-02 1 ## Mid1 1.561786e-06 0.5503712 0.188 0.085 6.232308e-02 1 ## Znhit3 1.571061e-06 0.7042544 0.294 0.160 6.269321e-02 1 ## Fam210b 1.578007e-06 0.3063875 0.339 0.191 6.297037e-02 1 ## Nipa1 1.613612e-06 0.2188123 0.155 0.061 6.439117e-02 1 ## Speer4c1 1.617908e-06 2.3175696 0.053 0.010 6.456262e-02 1 ## Gorasp1 1.635346e-06 0.6997876 0.208 0.099 6.525849e-02 1 ## Ergic11 1.636768e-06 0.4212578 0.514 0.320 6.531522e-02 1 ## Ube2q2 1.637036e-06 0.4380788 0.465 0.286 6.532594e-02 1 ## E2f5 1.643638e-06 0.5405930 0.343 0.198 6.558938e-02 1 ## Tmem2231 1.643771e-06 0.2038465 0.824 0.542 6.559470e-02 1 ## Kptn 1.651352e-06 0.6900082 0.290 0.154 6.589720e-02 1 ## Rtn21 1.652412e-06 0.3718708 0.494 0.293 6.593952e-02 1 ## Phlda2 1.659983e-06 2.0257697 0.053 0.010 6.624164e-02 1 ## Smpd1 1.685588e-06 0.4925625 0.347 0.191 6.726340e-02 1 ## Ficd 1.695963e-06 0.2912818 0.278 0.140 6.767739e-02 1 ## Capns11 1.704389e-06 0.1745488 0.939 0.702 6.801364e-02 1 ## Prrx21 1.709582e-06 0.3103053 0.400 0.230 6.822087e-02 1 ## Inpp5e 1.710539e-06 0.6317556 0.208 0.099 6.825906e-02 1 ## Dpp81 1.719322e-06 0.1670313 0.767 0.518 6.860955e-02 1 ## Slc35b21 1.747290e-06 0.2507186 0.559 0.345 6.972560e-02 1 ## Zbtb33 1.762368e-06 0.2875696 0.424 0.242 7.032730e-02 1 ## Slc16a12 1.764134e-06 1.8587570 0.078 0.022 7.039776e-02 1 ## Anxa7 1.781768e-06 0.3568960 0.576 0.362 7.110144e-02 1 ## Prmt21 1.786432e-06 0.2922390 0.547 0.333 7.128756e-02 1 ## Ube2r21 1.805356e-06 0.2090658 0.927 0.690 7.204273e-02 1 ## Ecscr 1.808926e-06 2.6623740 0.065 0.016 7.218520e-02 1 ## Tsr31 1.825560e-06 0.3681146 0.429 0.261 7.284897e-02 1 ## Olah 1.841884e-06 3.5834752 0.020 0.001 7.350040e-02 1 ## C1ql4 1.841884e-06 3.5954823 0.020 0.001 7.350040e-02 1 ## Gm4265 1.854554e-06 3.5711718 0.020 0.001 7.400597e-02 1 ## Insc 1.864530e-06 0.9057799 0.184 0.084 7.440408e-02 1 ## Hpcal4 1.867307e-06 2.8287764 0.020 0.001 7.451489e-02 1 ## Gm31889 1.867307e-06 3.2099399 0.020 0.001 7.451489e-02 1 ## LOC118568708 1.879083e-06 0.2867782 0.400 0.227 7.498480e-02 1 ## Nifk 1.882308e-06 0.2123788 0.461 0.277 7.511350e-02 1 ## Pigk 1.888954e-06 0.4139448 0.506 0.324 7.537870e-02 1 ## Ercc5 1.909003e-06 0.3630489 0.294 0.157 7.617878e-02 1 ## Tmem1601 1.944764e-06 0.1857578 0.902 0.644 7.760579e-02 1 ## Mark3 1.946293e-06 0.1912058 0.841 0.614 7.766682e-02 1 ## Ice1 1.948466e-06 0.3574806 0.408 0.238 7.775354e-02 1 ## Fank1 1.971700e-06 1.2965938 0.086 0.025 7.868067e-02 1 ## Ssu721 1.974231e-06 0.2270321 0.873 0.614 7.878169e-02 1 ## Nit1 1.995932e-06 0.2819307 0.608 0.400 7.964765e-02 1 ## Acap21 2.010390e-06 0.1532217 0.637 0.419 8.022460e-02 1 ## Gm44148 2.024649e-06 0.8761613 0.131 0.049 8.079363e-02 1 ## Parl 2.034868e-06 0.2207957 0.612 0.391 8.120142e-02 1 ## Armh4 2.041800e-06 3.1971124 0.037 0.005 8.147805e-02 1 ## Oxld1 2.082765e-06 0.5762942 0.269 0.147 8.311274e-02 1 ## Rad1 2.089592e-06 0.6636559 0.224 0.108 8.338518e-02 1 ## Atp6v0b1 2.095659e-06 0.1756658 0.992 0.857 8.362726e-02 1 ## Bbc3 2.101438e-06 0.2867723 0.339 0.187 8.385790e-02 1 ## Tle51 2.108204e-06 0.1775549 1.000 0.894 8.412790e-02 1 ## Gm53915 2.111765e-06 0.4457762 0.069 0.017 8.426998e-02 1 ## Cnep1r11 2.121535e-06 0.2501852 0.604 0.403 8.465987e-02 1 ## Morf4l1b1 2.149447e-06 0.4321550 0.478 0.303 8.577368e-02 1 ## Maip1 2.170722e-06 0.3423875 0.371 0.212 8.662264e-02 1 ## Dok41 2.171752e-06 0.4906470 0.551 0.371 8.666377e-02 1 ## 2510022D24Rik1 2.178252e-06 0.2816250 0.690 0.464 8.692313e-02 1 ## Dync1li2 2.209665e-06 0.1170325 0.682 0.456 8.817666e-02 1 ## Gm57842 2.210140e-06 2.1620736 0.065 0.016 8.819563e-02 1 ## P4ha11 2.215155e-06 0.2028430 0.412 0.233 8.839577e-02 1 ## 5033403F01Rik 2.219954e-06 2.0502618 0.049 0.009 8.858727e-02 1 ## Cfap144 2.222886e-06 0.1220775 0.980 0.746 8.870428e-02 1 ## Mettl1 2.235585e-06 0.2189345 0.310 0.160 8.921103e-02 1 ## Bmf 2.236606e-06 0.3540022 0.208 0.099 8.925175e-02 1 ## Ncbp21 2.241063e-06 0.3234785 0.498 0.299 8.942962e-02 1 ## Susd6 2.241261e-06 0.1177210 0.633 0.402 8.943752e-02 1 ## Exosc41 2.245619e-06 0.2896916 0.624 0.403 8.961144e-02 1 ## Gm26559 2.254060e-06 0.6035440 0.139 0.053 8.994825e-02 1 ## Bcl6 2.256957e-06 0.9903191 0.139 0.055 9.006387e-02 1 ## Gm36416 2.257563e-06 2.7151912 0.037 0.005 9.008805e-02 1 ## Dip2c 2.306863e-06 0.3822348 0.355 0.201 9.205537e-02 1 ## Ace31 2.353516e-06 0.8831999 0.188 0.085 9.391704e-02 1 ## Ccdc591 2.354143e-06 0.2725705 0.722 0.470 9.394208e-02 1 ## Atrnl11 2.365590e-06 0.2556255 0.469 0.280 9.439885e-02 1 ## Gca 2.373419e-06 0.3036306 0.331 0.188 9.471130e-02 1 ## Gm53314 2.393363e-06 2.3782439 0.037 0.005 9.550715e-02 1 ## Mllt6 2.404068e-06 0.2080331 0.482 0.290 9.593434e-02 1 ## Gm11655 2.407320e-06 2.5554655 0.041 0.006 9.606411e-02 1 ## 1810044D09Rik 2.410880e-06 0.8365818 0.204 0.099 9.620615e-02 1 ## Ppp1r3c 2.413392e-06 2.3335947 0.037 0.005 9.630641e-02 1 ## Mterf4 2.416575e-06 0.4439344 0.286 0.155 9.643341e-02 1 ## Cd200l11 2.421529e-06 0.1719348 0.947 0.734 9.663113e-02 1 ## Atg5 2.427558e-06 0.1222480 0.649 0.408 9.687171e-02 1 ## Mrpl50 2.443828e-06 0.3275254 0.518 0.317 9.752097e-02 1 ## Evi5 2.459334e-06 0.3973561 0.535 0.342 9.813973e-02 1 ## Tmem131l 2.473450e-06 0.3931332 0.400 0.237 9.870302e-02 1 ## Coro7 2.474505e-06 0.1522124 0.261 0.133 9.874512e-02 1 ## Nsa2 2.505048e-06 0.1617273 0.971 0.741 9.996396e-02 1 ## Sall4 2.505531e-06 2.1730395 0.037 0.005 9.998320e-02 1 ## Rnf61 2.513818e-06 0.1284173 0.873 0.645 1.003139e-01 1 ## Vasp1 2.523033e-06 0.1032200 0.931 0.692 1.006816e-01 1 ## Usp37 2.541930e-06 0.2286756 0.355 0.194 1.014357e-01 1 ## Rnf1411 2.546226e-06 0.1907766 0.727 0.505 1.016072e-01 1 ## Particl 2.554870e-06 0.8595861 0.131 0.051 1.019521e-01 1 ## Mllt111 2.562708e-06 0.7075172 0.261 0.136 1.022649e-01 1 ## Heatr5b 2.578518e-06 0.6420664 0.273 0.143 1.028958e-01 1 ## Bet1 2.586050e-06 0.1875130 0.555 0.342 1.031963e-01 1 ## Ints11 2.600938e-06 0.2499526 0.384 0.220 1.037904e-01 1 ## Stx6 2.606362e-06 0.5087388 0.290 0.155 1.040069e-01 1 ## Ube2n1 2.630730e-06 0.2470100 0.869 0.628 1.049793e-01 1 ## Traf5 2.663873e-06 1.3304317 0.090 0.028 1.063018e-01 1 ## Tmem86b1 2.685325e-06 0.1024774 0.722 0.475 1.071579e-01 1 ## Zc3h14 2.690457e-06 0.2411992 0.694 0.454 1.073627e-01 1 ## Lrrc24 2.711247e-06 0.5758703 0.188 0.085 1.081923e-01 1 ## Nab1 2.728101e-06 0.3820159 0.420 0.252 1.088649e-01 1 ## 4632428C04Rik 2.739685e-06 1.4420503 0.065 0.016 1.093271e-01 1 ## Gm30084 2.740916e-06 1.9527359 0.041 0.006 1.093763e-01 1 ## Zfp821 2.760411e-06 0.2405629 0.355 0.198 1.101542e-01 1 ## Egln11 2.789294e-06 0.1804301 0.612 0.393 1.113068e-01 1 ## Ccdc88c 2.815927e-06 0.4271167 0.314 0.177 1.123696e-01 1 ## Noa1 2.836779e-06 0.6547856 0.265 0.141 1.132017e-01 1 ## Pycr3 2.843299e-06 0.1587775 0.494 0.284 1.134618e-01 1 ## 1110019D14Rik 2.857740e-06 0.2299457 0.147 0.061 1.140381e-01 1 ## Dcaf6 2.877180e-06 0.1540517 0.416 0.241 1.148139e-01 1 ## Got2 2.889647e-06 0.3562697 0.547 0.343 1.153114e-01 1 ## Grn1 2.896128e-06 0.2132699 0.682 0.449 1.155700e-01 1 ## Zfp955b 2.896234e-06 0.1566966 0.318 0.173 1.155742e-01 1 ## Bbs5 2.898856e-06 0.9604075 0.118 0.043 1.156788e-01 1 ## Rtf2 2.933709e-06 0.2198310 0.682 0.439 1.170696e-01 1 ## Zcchc171 2.954202e-06 0.2727466 0.653 0.416 1.178874e-01 1 ## R3hdm41 2.966277e-06 0.2208507 0.780 0.530 1.183693e-01 1 ## Arfip11 2.983431e-06 0.1129855 0.600 0.404 1.190538e-01 1 ## Ints10 2.984009e-06 0.3701686 0.367 0.210 1.190769e-01 1 ## Lgmn 3.017549e-06 0.3358647 0.404 0.240 1.204153e-01 1 ## 1110059G10Rik 3.019054e-06 0.3324827 0.494 0.301 1.204754e-01 1 ## Ssr31 3.030146e-06 0.2242994 0.902 0.667 1.209180e-01 1 ## Zc2hc1a 3.038003e-06 0.2298520 0.327 0.176 1.212315e-01 1 ## Dtwd1 3.067200e-06 0.4424270 0.171 0.078 1.223966e-01 1 ## Dgcr2 3.081340e-06 0.3385357 0.543 0.342 1.229609e-01 1 ## Zbtb16 3.107026e-06 0.8977466 0.155 0.066 1.239859e-01 1 ## Gm26588 3.125419e-06 1.0949180 0.090 0.028 1.247199e-01 1 ## Zbtb41 3.157546e-06 0.3584117 0.424 0.266 1.260019e-01 1 ## Col8a1 3.170158e-06 0.3384294 0.208 0.099 1.265052e-01 1 ## Mfap31 3.193545e-06 0.1571567 0.510 0.310 1.274384e-01 1 ## Triqk1 3.195863e-06 0.2975070 0.376 0.210 1.275309e-01 1 ## Slc49a4 3.199614e-06 0.3197412 0.347 0.198 1.276806e-01 1 ## Atp2c11 3.215369e-06 0.1901486 0.714 0.526 1.283093e-01 1 ## Lman2l 3.216157e-06 0.4606973 0.298 0.164 1.283408e-01 1 ## Cbln31 3.273114e-06 0.4799542 0.208 0.096 1.306136e-01 1 ## Cog5 3.294510e-06 0.1706274 0.547 0.338 1.314674e-01 1 ## Uba7 3.313110e-06 0.2660510 0.310 0.169 1.322097e-01 1 ## Mindy11 3.330961e-06 0.1565188 0.624 0.402 1.329220e-01 1 ## Gm30871 3.336427e-06 1.0217242 0.082 0.024 1.331401e-01 1 ## Vamp81 3.431503e-06 0.2060522 0.976 0.790 1.369341e-01 1 ## Polr1h1 3.445041e-06 0.2724243 0.588 0.395 1.374744e-01 1 ## Zkscan8 3.446898e-06 0.3706088 0.294 0.159 1.375485e-01 1 ## Lama5 3.485421e-06 1.8376735 0.078 0.022 1.390857e-01 1 ## Cpt1c 3.497718e-06 0.2254564 0.180 0.078 1.395764e-01 1 ## Fuom 3.510414e-06 0.2322659 0.559 0.345 1.400831e-01 1 ## Grsf11 3.531924e-06 0.4002234 0.551 0.357 1.409414e-01 1 ## Jmjd8 3.567791e-06 0.4613937 0.400 0.239 1.423727e-01 1 ## 1110038F14Rik 3.577814e-06 0.1702345 0.633 0.415 1.427727e-01 1 ## Ufl1 3.632786e-06 0.1652194 0.686 0.456 1.449663e-01 1 ## Ttc1 3.635843e-06 0.1744810 0.522 0.325 1.450883e-01 1 ## Kdm7a1 3.650116e-06 0.1574633 0.931 0.702 1.456579e-01 1 ## Tmem231 3.670856e-06 1.5227504 0.073 0.020 1.464855e-01 1 ## Mnat1 3.686368e-06 0.4336347 0.339 0.194 1.471045e-01 1 ## Tmem167 3.739429e-06 0.1869324 0.865 0.624 1.492219e-01 1 ## Wbp21 3.760412e-06 0.1874724 0.849 0.584 1.500592e-01 1 ## Acot13 3.772055e-06 0.2470228 0.563 0.363 1.505239e-01 1 ## Coprs1 3.786475e-06 0.3258852 0.498 0.322 1.510993e-01 1 ## Tor2a 3.794457e-06 0.3116642 0.543 0.335 1.514178e-01 1 ## Ipo4 3.796129e-06 0.4472285 0.233 0.117 1.514845e-01 1 ## Gpn3 3.825791e-06 0.6744648 0.306 0.173 1.526682e-01 1 ## Vkorc1l11 3.832554e-06 0.2727401 0.445 0.271 1.529381e-01 1 ## Gm36180 3.891971e-06 3.5175146 0.029 0.003 1.553091e-01 1 ## Gm53213 3.894020e-06 1.4498658 0.086 0.027 1.553909e-01 1 ## Tmem175 3.925442e-06 0.6559024 0.245 0.129 1.566448e-01 1 ## Mapk1ip1 3.931609e-06 0.1142400 0.359 0.203 1.568909e-01 1 ## Gm36026 3.935150e-06 0.9464962 0.090 0.028 1.570322e-01 1 ## Nln 3.951292e-06 0.6666632 0.282 0.152 1.576763e-01 1 ## Gm458711 3.976485e-06 0.1605684 0.514 0.317 1.586816e-01 1 ## Psmd31 3.987407e-06 0.1345172 0.714 0.472 1.591175e-01 1 ## Gm46915 3.988859e-06 2.5293341 0.029 0.003 1.591754e-01 1 ## Ambp1 4.016880e-06 0.1298674 0.894 0.725 1.602936e-01 1 ## Khdrbs3 4.034943e-06 0.2738409 0.547 0.338 1.610144e-01 1 ## Tcea2 4.056611e-06 1.9820316 0.053 0.011 1.618791e-01 1 ## Nectin4 4.068029e-06 2.8537423 0.029 0.003 1.623347e-01 1 ## Cdc371 4.098049e-06 0.2527959 0.886 0.640 1.635326e-01 1 ## Pycr1 4.133683e-06 1.8143598 0.065 0.017 1.649546e-01 1 ## Ift88 4.185610e-06 0.2380890 0.265 0.138 1.670268e-01 1 ## Zfyve27 4.186475e-06 0.2408924 0.449 0.272 1.670613e-01 1 ## 0610009E02Rik 4.242078e-06 0.8135614 0.135 0.053 1.692801e-01 1 ## Impa11 4.258525e-06 0.2030890 0.861 0.620 1.699365e-01 1 ## Urod 4.274013e-06 0.3677814 0.461 0.287 1.705545e-01 1 ## Jade31 4.448943e-06 0.2578776 0.453 0.276 1.775351e-01 1 ## Coa6 4.451655e-06 0.2272236 0.518 0.319 1.776433e-01 1 ## Ptk71 4.464711e-06 0.6782516 0.163 0.070 1.781643e-01 1 ## Kcnd1 4.542115e-06 0.2911106 0.167 0.073 1.812531e-01 1 ## Abhd17b 4.557759e-06 0.2663859 0.584 0.378 1.818774e-01 1 ## Mid1ip11 4.690986e-06 0.1204693 0.922 0.699 1.871938e-01 1 ## Mesd 4.740346e-06 0.1597252 0.518 0.308 1.891635e-01 1 ## Zfp51 4.783173e-06 0.6506044 0.229 0.115 1.908725e-01 1 ## Edc3 4.791082e-06 0.4880992 0.282 0.150 1.911881e-01 1 ## A430046D13Rik 4.888223e-06 1.1027775 0.086 0.027 1.950645e-01 1 ## Ptcra 4.913590e-06 0.9912259 0.122 0.047 1.960768e-01 1 ## Slc25a13 5.034072e-06 0.1436755 0.927 0.667 2.008846e-01 1 ## Usp19 5.071350e-06 0.1257201 0.714 0.477 2.023722e-01 1 ## Gnl3l1 5.077429e-06 0.1646133 0.727 0.505 2.026148e-01 1 ## Gm5113 5.084087e-06 0.7750746 0.143 0.058 2.028805e-01 1 ## Dusp7 5.165359e-06 1.4827445 0.086 0.027 2.061237e-01 1 ## Hspa51 5.186139e-06 0.1446050 0.988 0.890 2.069529e-01 1 ## Ttc22 5.198150e-06 0.2806617 0.371 0.215 2.074322e-01 1 ## Mterf2 5.198219e-06 0.2422943 0.335 0.190 2.074349e-01 1 ## Itga3 5.225324e-06 0.1006629 0.400 0.233 2.085165e-01 1 ## Ubxn10 5.334712e-06 0.5547338 0.196 0.093 2.128817e-01 1 ## 2310057M21Rik 5.340118e-06 0.3650960 0.220 0.110 2.130974e-01 1 ## Ube2w1 5.359061e-06 0.2531742 0.633 0.430 2.138533e-01 1 ## Cnpy4 5.360741e-06 0.5281981 0.282 0.152 2.139204e-01 1 ## Gm17753 5.381073e-06 0.7663776 0.151 0.064 2.147317e-01 1 ## Ccdc171 5.415156e-06 0.8341579 0.110 0.041 2.160918e-01 1 ## Wdr47 5.429649e-06 0.1742631 0.298 0.154 2.166702e-01 1 ## Tasp1 5.517645e-06 0.5475920 0.200 0.094 2.201816e-01 1 ## B330016D10Rik 5.576241e-06 0.2771499 0.290 0.155 2.225199e-01 1 ## Nova2 5.586712e-06 0.5195995 0.192 0.088 2.229377e-01 1 ## Cryzl1 5.643284e-06 0.3098947 0.571 0.365 2.251953e-01 1 ## Gm31356 5.676797e-06 1.1560570 0.065 0.017 2.265326e-01 1 ## Mgat4b 5.679126e-06 0.1815415 0.661 0.431 2.266255e-01 1 ## Pfdn41 5.753182e-06 0.3906966 0.469 0.293 2.295807e-01 1 ## Mrpl551 5.757848e-06 0.1056902 0.547 0.328 2.297669e-01 1 ## Ric8a 5.760030e-06 0.1407335 0.547 0.344 2.298540e-01 1 ## H2aj1 5.817251e-06 0.1704482 0.996 0.827 2.321374e-01 1 ## Gar1 5.836504e-06 0.2618265 0.253 0.127 2.329057e-01 1 ## Eif4ebp1 5.853407e-06 0.7796323 0.249 0.131 2.335802e-01 1 ## Trim13 5.865639e-06 1.1521394 0.122 0.048 2.340683e-01 1 ## G3bp21 5.866501e-06 0.1541136 0.922 0.697 2.341027e-01 1 ## Chst15 5.871918e-06 1.2943338 0.045 0.008 2.343189e-01 1 ## Ccdc90b 5.905296e-06 0.3716813 0.335 0.197 2.356508e-01 1 ## Cyp20a11 5.912121e-06 0.3522305 0.416 0.244 2.359232e-01 1 ## Lpcat1 5.929419e-06 0.4224174 0.253 0.129 2.366135e-01 1 ## Cdr2 5.948657e-06 0.3524594 0.245 0.127 2.373812e-01 1 ## Ykt61 5.949855e-06 0.1202345 0.682 0.484 2.374290e-01 1 ## Afap1 5.958055e-06 0.7322488 0.171 0.078 2.377562e-01 1 ## Mpg 6.023791e-06 0.4570887 0.310 0.175 2.403794e-01 1 ## Mettl25 6.037678e-06 0.6035055 0.233 0.119 2.409335e-01 1 ## Sdr39u1 6.089796e-06 0.5577495 0.253 0.134 2.430133e-01 1 ## Ndufb8 6.104795e-06 0.1199383 0.959 0.729 2.436118e-01 1 ## Zfp523 6.184278e-06 0.3265809 0.306 0.173 2.467836e-01 1 ## Rnf21 6.228038e-06 0.2984704 0.408 0.235 2.485298e-01 1 ## Dr11 6.229313e-06 0.3715316 0.490 0.308 2.485807e-01 1 ## Tdrd3 6.268148e-06 0.2134474 0.445 0.279 2.501305e-01 1 ## Apoa2 6.268577e-06 0.9094565 0.147 0.062 2.501476e-01 1 ## Gm32164 6.319418e-06 2.0574341 0.033 0.004 2.521764e-01 1 ## Gm35574 6.346500e-06 2.5232004 0.033 0.004 2.532571e-01 1 ## Atxn3 6.371981e-06 0.2113580 0.469 0.289 2.542739e-01 1 ## Mrps28 6.407561e-06 0.3328307 0.441 0.256 2.556937e-01 1 ## Ppp3cc 6.407749e-06 0.3113536 0.212 0.103 2.557012e-01 1 ## Prkch 6.411580e-06 2.7627732 0.041 0.007 2.558541e-01 1 ## Akr1b10 6.467780e-06 0.4180312 0.094 0.032 2.580968e-01 1 ## Cuedc21 6.480872e-06 0.2644308 0.653 0.427 2.586192e-01 1 ## Alad 6.493933e-06 0.3023936 0.506 0.310 2.591404e-01 1 ## Ddx421 6.537151e-06 0.2694407 0.743 0.526 2.608650e-01 1 ## Tmx21 6.555863e-06 0.2961654 0.498 0.308 2.616117e-01 1 ## Gm9902 6.577100e-06 0.8106886 0.122 0.048 2.624592e-01 1 ## Cers51 6.595728e-06 0.3317725 0.441 0.267 2.632025e-01 1 ## Zfp788 6.612713e-06 0.3517836 0.245 0.124 2.638803e-01 1 ## Dnajb3 6.621162e-06 1.4568746 0.082 0.025 2.642175e-01 1 ## Pip4k2b 6.621953e-06 0.6056642 0.184 0.086 2.642490e-01 1 ## Cstad 6.651824e-06 2.1156004 0.033 0.004 2.654410e-01 1 ## Med10 6.661273e-06 0.2303820 0.424 0.255 2.658181e-01 1 ## Txlna 6.680775e-06 0.2246691 0.518 0.329 2.665963e-01 1 ## Cbx8 6.685590e-06 0.5694751 0.212 0.103 2.667885e-01 1 ## Rhbdl1 6.712633e-06 0.6861673 0.167 0.075 2.678676e-01 1 ## Snhg12 6.740364e-06 0.3786632 0.302 0.174 2.689742e-01 1 ## Gtf3c61 6.789771e-06 0.1797778 0.576 0.356 2.709458e-01 1 ## Map4k4 6.796078e-06 0.1541433 0.784 0.578 2.711975e-01 1 ## Hpn 6.834762e-06 2.4062623 0.053 0.012 2.727412e-01 1 ## Mrpl111 6.869865e-06 0.2130960 0.649 0.426 2.741420e-01 1 ## Lsm7 6.906658e-06 0.1287523 0.567 0.347 2.756102e-01 1 ## Akap111 6.914683e-06 0.1537697 0.718 0.484 2.759304e-01 1 ## Glod41 6.962780e-06 0.2315033 0.767 0.516 2.778497e-01 1 ## Stt3a1 7.013903e-06 0.2246436 0.804 0.572 2.798898e-01 1 ## Kantr 7.052241e-06 0.3794856 0.273 0.148 2.814197e-01 1 ## Stox2 7.238422e-06 1.5787590 0.090 0.030 2.888492e-01 1 ## Gjb11 7.253243e-06 0.3665604 0.331 0.187 2.894406e-01 1 ## Dph7 7.399563e-06 1.1792674 0.106 0.038 2.952796e-01 1 ## Cbarp 7.404283e-06 0.3098585 0.388 0.224 2.954679e-01 1 ## Slc38a10 7.497936e-06 0.2059289 0.710 0.465 2.992051e-01 1 ## Rit1 7.515291e-06 0.2370848 0.502 0.317 2.998977e-01 1 ## Rubcn 7.622088e-06 0.3652060 0.294 0.166 3.041594e-01 1 ## Skic81 7.660974e-06 0.1656733 0.710 0.459 3.057112e-01 1 ## Arf11 7.692919e-06 0.2113619 0.988 0.827 3.069859e-01 1 ## Rab5b1 7.713333e-06 0.1525698 0.624 0.407 3.078006e-01 1 ## Rbm101 7.782123e-06 0.3044527 0.645 0.438 3.105456e-01 1 ## Zfp280c 7.795762e-06 0.4630497 0.196 0.094 3.110899e-01 1 ## Tor1a 7.806376e-06 0.2938959 0.502 0.321 3.115134e-01 1 ## Eif4a11 7.809311e-06 0.1313478 1.000 0.813 3.116306e-01 1 ## Fam131c 7.832058e-06 0.8763140 0.082 0.025 3.125383e-01 1 ## Hopxos 7.860443e-06 0.4237829 0.102 0.036 3.136710e-01 1 ## Magohb 7.893261e-06 0.3490577 0.294 0.161 3.149806e-01 1 ## Pla2g12b 7.994486e-06 0.2307308 0.518 0.335 3.190200e-01 1 ## Gramd4 8.043084e-06 0.4008322 0.233 0.122 3.209593e-01 1 ## Tent5a 8.099979e-06 0.1430031 0.955 0.802 3.232296e-01 1 ## Slc35a1 8.126045e-06 0.3126003 0.539 0.360 3.242698e-01 1 ## Brip1os 8.150475e-06 0.2934067 0.457 0.277 3.252447e-01 1 ## Gpc4 8.175784e-06 0.1633839 0.637 0.391 3.262547e-01 1 ## Sec23b 8.262619e-06 0.1515447 0.661 0.456 3.297198e-01 1 ## Nutf21 8.321188e-06 0.1572988 0.698 0.463 3.320570e-01 1 ## Snrpb21 8.375646e-06 0.2029970 0.498 0.305 3.342301e-01 1 ## Bckdk 8.453753e-06 0.4030664 0.449 0.277 3.373470e-01 1 ## Tusc2 8.473580e-06 0.2887634 0.494 0.317 3.381382e-01 1 ## Cep120 8.474221e-06 0.1125886 0.331 0.180 3.381638e-01 1 ## Psmg1 8.592115e-06 0.2435017 0.306 0.171 3.428683e-01 1 ## Snx31 8.609115e-06 0.1632793 0.963 0.779 3.435468e-01 1 ## Smurf1 8.633874e-06 0.3532772 0.298 0.167 3.445348e-01 1 ## Tgs1 8.640738e-06 0.1096419 0.539 0.341 3.448086e-01 1 ## Gm42250 8.651908e-06 0.6590010 0.053 0.012 3.452544e-01 1 ## Ube2l31 8.835927e-06 0.1972566 0.927 0.689 3.525977e-01 1 ## Cyb5d1 8.847281e-06 0.6689552 0.229 0.118 3.530508e-01 1 ## Dhdds 8.860551e-06 0.2732274 0.433 0.265 3.535803e-01 1 ## Mob3a 8.878143e-06 0.5147598 0.212 0.106 3.542823e-01 1 ## Xpo7 9.009012e-06 0.2467251 0.514 0.326 3.595046e-01 1 ## Rgp1 9.015382e-06 0.3279885 0.331 0.192 3.597588e-01 1 ## Bub31 9.043808e-06 0.1905628 0.645 0.428 3.608931e-01 1 ## Maneal 9.072737e-06 1.0107819 0.127 0.051 3.620476e-01 1 ## B230219D22Rik1 9.087260e-06 0.2231031 0.882 0.667 3.626271e-01 1 ## Ppil21 9.116012e-06 0.1370485 0.637 0.430 3.637744e-01 1 ## Pou3f4 9.140743e-06 4.3101042 0.024 0.002 3.647614e-01 1 ## Ankrd39 9.204680e-06 0.8303943 0.184 0.090 3.673127e-01 1 ## Srd5a3 9.230403e-06 0.4787358 0.314 0.177 3.683392e-01 1 ## Gm32031 9.242788e-06 0.4284768 0.257 0.134 3.688335e-01 1 ## Pnpla6 9.268083e-06 0.1190483 0.539 0.342 3.698429e-01 1 ## Atpsckmt 9.292232e-06 0.2857641 0.347 0.203 3.708065e-01 1 ## Tmem109 9.351753e-06 0.3249024 0.351 0.204 3.731817e-01 1 ## Ddx31 9.386732e-06 0.6610801 0.143 0.061 3.745776e-01 1 ## Mettl4 9.430011e-06 0.2298757 0.318 0.175 3.763046e-01 1 ## Cyb5611 9.438231e-06 0.1267923 0.763 0.518 3.766326e-01 1 ## Exosc5 9.472315e-06 0.2058794 0.457 0.280 3.779927e-01 1 ## Mbd4 9.479171e-06 1.0010420 0.163 0.075 3.782663e-01 1 ## 1700040D17Rik 9.482003e-06 3.2386222 0.024 0.002 3.783793e-01 1 ## A930041C12Rik 9.482003e-06 3.1555015 0.024 0.002 3.783793e-01 1 ## Fez1 9.482003e-06 3.0749688 0.024 0.002 3.783793e-01 1 ## Uprt1 9.550511e-06 0.3300714 0.380 0.223 3.811131e-01 1 ## Pcif11 9.564685e-06 0.2212335 0.616 0.412 3.816787e-01 1 ## Capza21 9.603701e-06 0.1774016 0.927 0.704 3.832357e-01 1 ## Ufd1 9.677534e-06 0.2702807 0.514 0.331 3.861820e-01 1 ## Scoc1 9.719194e-06 0.2293178 0.608 0.409 3.878444e-01 1 ## Gm29415 9.733176e-06 1.9671106 0.024 0.002 3.884024e-01 1 ## Tead41 9.808094e-06 0.8046503 0.098 0.034 3.913920e-01 1 ## Hsd17b8 9.839667e-06 0.2719696 0.486 0.313 3.926519e-01 1 ## Pam161 9.970132e-06 0.1814165 0.894 0.633 3.978581e-01 1 ## Pgm3 9.971892e-06 0.1762920 0.490 0.307 3.979283e-01 1 ## Gm54265 1.006528e-05 1.8939017 0.061 0.016 4.016551e-01 1 ## Ier3ip11 1.015996e-05 0.1642751 0.959 0.714 4.054331e-01 1 ## Gm39469 1.017177e-05 0.6487049 0.167 0.076 4.059043e-01 1 ## Ptch1 1.026866e-05 0.2158596 0.367 0.218 4.097711e-01 1 ## Lhpp 1.028848e-05 0.6092301 0.212 0.109 4.105620e-01 1 ## Pip4p1 1.030634e-05 0.2187419 0.555 0.341 4.112744e-01 1 ## Anapc4 1.030891e-05 0.2596754 0.482 0.303 4.113770e-01 1 ## Rnf13 1.035495e-05 0.1762259 0.506 0.312 4.132143e-01 1 ## Gemin2 1.047290e-05 0.7333447 0.224 0.118 4.179209e-01 1 ## Chpf2 1.051835e-05 0.2642966 0.278 0.149 4.197349e-01 1 ## Gm38872 1.053012e-05 0.7843684 0.155 0.068 4.202045e-01 1 ## Ccnq 1.054509e-05 0.3002986 0.314 0.179 4.208019e-01 1 ## Arap1 1.056107e-05 0.1785306 0.429 0.262 4.214394e-01 1 ## Jakmip11 1.061919e-05 0.3218711 0.482 0.312 4.237586e-01 1 ## Npr11 1.064189e-05 0.9734319 0.159 0.072 4.246647e-01 1 ## Dcaf71 1.090048e-05 0.1888529 0.665 0.465 4.349836e-01 1 ## Cradd 1.096718e-05 0.1341939 0.335 0.194 4.376454e-01 1 ## Srrm3 1.104916e-05 0.1674816 0.367 0.208 4.409168e-01 1 ## 4933434E20Rik 1.110595e-05 0.3822205 0.433 0.266 4.431830e-01 1 ## Yif1a1 1.112014e-05 0.1951250 0.682 0.452 4.437493e-01 1 ## Cfap119 1.113867e-05 1.0464463 0.102 0.037 4.444885e-01 1 ## Dnal4 1.114265e-05 0.5578445 0.233 0.124 4.446474e-01 1 ## Fbxo8 1.116722e-05 0.2456536 0.502 0.311 4.456281e-01 1 ## Rala1 1.118735e-05 0.1816719 0.939 0.699 4.464313e-01 1 ## Tpcn1 1.119461e-05 0.1138642 0.302 0.168 4.467207e-01 1 ## Gm53613 1.120566e-05 1.6995521 0.061 0.016 4.471618e-01 1 ## 4933421O10Rik 1.125047e-05 0.5935930 0.282 0.157 4.489498e-01 1 ## Phka2 1.133395e-05 0.4304737 0.257 0.139 4.522811e-01 1 ## Ccm2 1.142915e-05 0.3501247 0.294 0.165 4.560803e-01 1 ## Srprb 1.145794e-05 0.4394982 0.433 0.268 4.572291e-01 1 ## Sf3b31 1.150189e-05 0.2580892 0.559 0.358 4.589831e-01 1 ## Cdk5 1.158420e-05 0.1353620 0.461 0.287 4.622676e-01 1 ## Glis2 1.161464e-05 0.1515097 0.233 0.119 4.634824e-01 1 ## Gins4 1.166039e-05 0.5104207 0.286 0.163 4.653078e-01 1 ## Gpam 1.166664e-05 0.9363847 0.163 0.075 4.655573e-01 1 ## Dnajc17 1.170915e-05 0.6536721 0.233 0.124 4.672538e-01 1 ## Card6 1.173531e-05 0.4250742 0.224 0.114 4.682974e-01 1 ## Malsu1 1.176031e-05 0.3213814 0.531 0.335 4.692952e-01 1 ## Mrto4 1.178942e-05 0.3489779 0.306 0.166 4.704569e-01 1 ## Tmed91 1.179083e-05 0.1155117 0.780 0.521 4.705131e-01 1 ## Zfp513 1.187590e-05 0.1278966 0.310 0.171 4.739077e-01 1 ## Nat8f1 1.189597e-05 0.4041545 0.290 0.161 4.747088e-01 1 ## Psmb71 1.190317e-05 0.2034094 0.816 0.572 4.749961e-01 1 ## Msl11 1.194979e-05 0.1562849 0.804 0.577 4.768565e-01 1 ## Yaf2 1.203188e-05 0.2145285 0.396 0.237 4.801322e-01 1 ## Eif3e 1.209951e-05 0.1277268 0.918 0.632 4.828310e-01 1 ## Senp8 1.211900e-05 0.7974310 0.212 0.110 4.836088e-01 1 ## Gm348021 1.216418e-05 0.8015311 0.102 0.036 4.854114e-01 1 ## Ralb 1.224794e-05 0.3462314 0.437 0.272 4.887540e-01 1 ## Zbtb42 1.230715e-05 0.1039071 0.698 0.474 4.911167e-01 1 ## Gm32483 1.234637e-05 0.3419894 0.245 0.127 4.926820e-01 1 ## Twsg1 1.238405e-05 0.1399889 0.673 0.455 4.941857e-01 1 ## D630003M21Rik 1.248056e-05 1.5355076 0.049 0.010 4.980368e-01 1 ## Foxk2 1.249291e-05 0.2657523 0.429 0.259 4.985295e-01 1 ## Pipox 1.252824e-05 0.1696567 0.584 0.372 4.999396e-01 1 ## Dennd5a 1.253263e-05 0.2786274 0.314 0.178 5.001144e-01 1 ## Zfp146 1.265214e-05 0.4722539 0.376 0.226 5.048835e-01 1 ## Ndfip11 1.270322e-05 0.1687030 0.988 0.797 5.069218e-01 1 ## Dnaaf10 1.293884e-05 0.4784829 0.269 0.152 5.163245e-01 1 ## Gm156581 1.296632e-05 0.2824048 0.273 0.146 5.174209e-01 1 ## B4galt31 1.300189e-05 0.2773728 0.576 0.393 5.188405e-01 1 ## Rpap3 1.306397e-05 0.4467460 0.273 0.151 5.213179e-01 1 ## Abcc6 1.306602e-05 1.7036573 0.053 0.013 5.213994e-01 1 ## Nmb1 1.307883e-05 0.7670435 0.139 0.058 5.219107e-01 1 ## Acbd31 1.312364e-05 0.1443242 0.878 0.637 5.236989e-01 1 ## Rab19 1.327821e-05 0.6221564 0.261 0.145 5.298670e-01 1 ## Entrep3 1.352679e-05 0.3516829 0.163 0.075 5.397865e-01 1 ## Cnp 1.357096e-05 0.9091958 0.241 0.132 5.415493e-01 1 ## 1700102J08Rik 1.360753e-05 2.0362456 0.045 0.009 5.430087e-01 1 ## AI987944 1.361133e-05 0.5368339 0.253 0.136 5.431603e-01 1 ## Fpgs 1.365248e-05 1.1040335 0.131 0.056 5.448023e-01 1 ## Uri1 1.365883e-05 0.3133032 0.506 0.327 5.450558e-01 1 ## Gm26205 1.378142e-05 0.5431178 0.204 0.103 5.499476e-01 1 ## Fyttd11 1.383184e-05 0.2253076 0.755 0.528 5.519597e-01 1 ## Rcor3 1.407943e-05 0.2899848 0.473 0.306 5.618397e-01 1 ## Gm32655 1.410816e-05 0.8542547 0.098 0.035 5.629862e-01 1 ## Denr1 1.415136e-05 0.1409673 0.869 0.606 5.647099e-01 1 ## Mta3 1.446732e-05 0.1576465 0.465 0.294 5.773182e-01 1 ## Dpm21 1.460119e-05 0.2656154 0.665 0.441 5.826607e-01 1 ## Cdk8 1.496533e-05 0.1113253 0.776 0.546 5.971915e-01 1 ## 4732491K20Rik 1.509342e-05 0.8140802 0.098 0.035 6.023031e-01 1 ## Gm49958 1.515017e-05 0.6326540 0.127 0.052 6.045676e-01 1 ## 5930430L01Rik 1.516743e-05 0.3006665 0.278 0.150 6.052563e-01 1 ## Gm57826 1.519995e-05 1.2473181 0.094 0.033 6.065539e-01 1 ## Bcap311 1.523233e-05 0.1636872 0.890 0.642 6.078459e-01 1 ## Sfrp5 1.530824e-05 0.2089105 0.269 0.148 6.108751e-01 1 ## Mogs 1.542658e-05 0.2353928 0.429 0.261 6.155977e-01 1 ## Mboat2 1.561481e-05 0.2051697 0.318 0.175 6.231090e-01 1 ## Cfap97 1.584622e-05 0.2032986 0.535 0.347 6.323434e-01 1 ## Tmem11 1.597192e-05 0.5437663 0.404 0.249 6.373595e-01 1 ## B3galnt2 1.597377e-05 0.3638445 0.278 0.152 6.374333e-01 1 ## Tor4a 1.602564e-05 0.3671172 0.261 0.140 6.395031e-01 1 ## Mplkip 1.610674e-05 0.2606268 0.510 0.325 6.427395e-01 1 ## Fbxo32 1.619053e-05 0.2420818 0.302 0.168 6.460833e-01 1 ## Kctd6 1.632303e-05 0.4030074 0.253 0.135 6.513704e-01 1 ## Cep164 1.672555e-05 0.1890659 0.620 0.398 6.674331e-01 1 ## Wdr11 1.690081e-05 0.2761268 0.420 0.261 6.744267e-01 1 ## Yipf51 1.691171e-05 0.1742330 0.739 0.514 6.748620e-01 1 ## Clk3 1.692842e-05 0.1442200 0.502 0.327 6.755284e-01 1 ## Ahi11 1.695224e-05 0.1219596 0.592 0.384 6.764790e-01 1 ## Zfp729b 1.708372e-05 0.3261559 0.261 0.140 6.817258e-01 1 ## Zfp977 1.719067e-05 2.6128710 0.029 0.003 6.859937e-01 1 ## Rtbdn 1.719067e-05 2.8083139 0.029 0.003 6.859937e-01 1 ## Rps6kl1 1.744810e-05 0.8795580 0.094 0.033 6.962665e-01 1 ## Cyp2c67 1.753247e-05 1.5547834 0.069 0.020 6.996332e-01 1 ## Oaz21 1.760152e-05 0.2323642 0.547 0.359 7.023888e-01 1 ## Borcs81 1.762673e-05 0.1267819 0.539 0.360 7.033948e-01 1 ## Capn7 1.765355e-05 0.1063234 0.547 0.360 7.044650e-01 1 ## Zbtb8b 1.766123e-05 2.0603287 0.041 0.008 7.047714e-01 1 ## Rybp1 1.777813e-05 0.2372946 0.433 0.259 7.094363e-01 1 ## Gm12216 1.782564e-05 0.2476515 0.294 0.164 7.113323e-01 1 ## Mbtps1 1.788976e-05 0.1120836 0.702 0.468 7.138909e-01 1 ## Stag2 1.802190e-05 0.1297327 0.686 0.452 7.191640e-01 1 ## Ppm1n 1.803607e-05 1.2215930 0.098 0.036 7.197293e-01 1 ## Mon1b 1.806534e-05 0.4867153 0.273 0.151 7.208973e-01 1 ## Srek1ip1 1.807234e-05 0.2119666 0.649 0.425 7.211768e-01 1 ## Dnajc221 1.809199e-05 0.3366819 0.543 0.369 7.219610e-01 1 ## Ppt2 1.809508e-05 0.5699294 0.196 0.099 7.220840e-01 1 ## Cdv3 1.809938e-05 0.1220023 0.796 0.544 7.222556e-01 1 ## Mrm2 1.827499e-05 0.7093197 0.216 0.114 7.292633e-01 1 ## Cacng4 1.832968e-05 0.7742091 0.098 0.036 7.314459e-01 1 ## Mrpl32 1.841441e-05 0.1365596 0.678 0.450 7.348268e-01 1 ## Il18bp 1.869721e-05 0.6322456 0.224 0.119 7.461122e-01 1 ## 4921531C22Rik 1.871233e-05 0.9538136 0.114 0.045 7.467156e-01 1 ## Fam219b 1.879194e-05 0.5071836 0.220 0.115 7.498924e-01 1 ## Josd21 1.881686e-05 0.2700516 0.555 0.365 7.508868e-01 1 ## Dhx58 1.889400e-05 0.8071239 0.106 0.041 7.539649e-01 1 ## Eif2b3 1.904984e-05 0.5175883 0.233 0.125 7.601838e-01 1 ## C2cd4c1 1.905277e-05 0.3525013 0.335 0.189 7.603006e-01 1 ## Tbc1d15 1.912714e-05 0.2003697 0.543 0.361 7.632685e-01 1 ## Psmd10 1.918127e-05 0.4073438 0.445 0.286 7.654285e-01 1 ## Timm9 1.919386e-05 0.2198863 0.347 0.199 7.659310e-01 1 ## Tbck 1.919416e-05 0.1251058 0.429 0.271 7.659430e-01 1 ## Dynlt1a 1.920554e-05 0.2215550 0.376 0.226 7.663971e-01 1 ## Elp4 1.923187e-05 0.3566627 0.302 0.173 7.674479e-01 1 ## Sacm1l1 1.928414e-05 0.1737831 0.641 0.434 7.695336e-01 1 ## 9530082P21Rik 1.938023e-05 0.6484761 0.135 0.057 7.733682e-01 1 ## Ranbp91 1.949417e-05 0.1991880 0.535 0.342 7.779149e-01 1 ## Zmat1 1.949833e-05 0.5430079 0.151 0.066 7.780808e-01 1 ## Ap1m21 1.970332e-05 0.2727628 0.629 0.413 7.862611e-01 1 ## Cope 1.970857e-05 0.1649628 0.890 0.688 7.864706e-01 1 ## Clstn3 1.974917e-05 2.4430795 0.033 0.005 7.880907e-01 1 ## Lrrc201 1.980968e-05 0.3540732 0.327 0.187 7.905053e-01 1 ## Ofd1 2.008307e-05 0.1305244 0.273 0.152 8.014151e-01 1 ## 2810029C07Rik 2.019540e-05 1.4598105 0.061 0.017 8.058976e-01 1 ## 2610306M01Rik1 2.021744e-05 0.3557714 0.306 0.171 8.067769e-01 1 ## Cdc16 2.030062e-05 0.3401131 0.416 0.253 8.100964e-01 1 ## Lrif1 2.037331e-05 0.2129178 0.563 0.382 8.129970e-01 1 ## Tapbp1 2.044425e-05 0.1973051 0.947 0.742 8.158278e-01 1 ## Crem 2.049068e-05 0.4476433 0.233 0.122 8.176805e-01 1 ## Hipk1 2.057198e-05 0.1502813 0.820 0.597 8.209247e-01 1 ## Gng7 2.071376e-05 1.9569084 0.037 0.006 8.265824e-01 1 ## Pnkp1 2.077014e-05 0.1566970 0.424 0.268 8.288326e-01 1 ## Gm54251 2.077722e-05 3.1228032 0.020 0.001 8.291151e-01 1 ## Gm32633 2.077722e-05 2.7282660 0.020 0.001 8.291151e-01 1 ## Ntm 2.078167e-05 2.2024016 0.033 0.005 8.292927e-01 1 ## Hebp2 2.079670e-05 0.7730849 0.180 0.087 8.298922e-01 1 ## Tbc1d23 2.084798e-05 0.2130213 0.457 0.295 8.319385e-01 1 ## Spag8 2.086248e-05 2.2690444 0.037 0.006 8.325174e-01 1 ## 1110028F18Rik 2.089635e-05 2.3000061 0.020 0.001 8.338687e-01 1 ## Crb31 2.131184e-05 0.2486700 0.714 0.490 8.504490e-01 1 ## Tbl1x1 2.138487e-05 0.1903391 0.829 0.602 8.533632e-01 1 ## Aldh5a1 2.148633e-05 0.3049091 0.265 0.147 8.574120e-01 1 ## Prima11 2.161181e-05 0.8170768 0.082 0.027 8.624193e-01 1 ## Gm12996 2.186544e-05 2.3280326 0.033 0.005 8.725405e-01 1 ## Cstb1 2.188093e-05 0.1738635 0.833 0.607 8.731585e-01 1 ## Zfp1481 2.207200e-05 0.1373081 0.886 0.666 8.807830e-01 1 ## Kctd7 2.210343e-05 0.6069222 0.167 0.078 8.820375e-01 1 ## Cltb1 2.234429e-05 0.1845987 0.841 0.618 8.916489e-01 1 ## Lamtor31 2.238824e-05 0.1581477 0.637 0.443 8.934026e-01 1 ## Glmp1 2.251668e-05 0.1918008 0.763 0.530 8.985282e-01 1 ## Cct6a1 2.276172e-05 0.2044177 0.873 0.637 9.083066e-01 1 ## Slc39a3 2.299829e-05 0.2068795 0.400 0.251 9.177470e-01 1 ## Gm14703 2.328956e-05 1.5100034 0.049 0.011 9.293697e-01 1 ## 9530062K07Rik 2.332319e-05 1.4861604 0.061 0.017 9.307119e-01 1 ## Actr31 2.345247e-05 0.1295603 0.971 0.818 9.358708e-01 1 ## Gm30842 2.355118e-05 0.5013991 0.135 0.057 9.398097e-01 1 ## Acvr1 2.365392e-05 0.5049411 0.359 0.222 9.439096e-01 1 ## Arl8a1 2.385839e-05 0.1659539 0.727 0.488 9.520689e-01 1 ## Adcy91 2.402150e-05 0.1452199 0.547 0.357 9.585778e-01 1 ## Klhl281 2.416394e-05 0.2532851 0.380 0.231 9.642620e-01 1 ## Zfp995 2.423444e-05 0.6289711 0.310 0.186 9.670755e-01 1 ## Sf3b61 2.428473e-05 0.1683024 0.959 0.725 9.690821e-01 1 ## Sumo31 2.436947e-05 0.1242293 0.796 0.559 9.724638e-01 1 ## Trim21 2.442624e-05 0.6572754 0.163 0.077 9.747290e-01 1 ## Cmc2 2.480696e-05 0.2491133 0.286 0.160 9.899216e-01 1 ## Scaper 2.480920e-05 0.1846389 0.482 0.318 9.900111e-01 1 ## Leprot1 2.510207e-05 0.1915806 0.600 0.409 1.000000e+00 1 ## 1110006O24Rik 2.527129e-05 1.1447406 0.102 0.039 1.000000e+00 1 ## Gm39526 2.564036e-05 1.3820755 0.069 0.021 1.000000e+00 1 ## Anapc7 2.585048e-05 0.3311775 0.376 0.226 1.000000e+00 1 ## Plppr21 2.587203e-05 0.3078864 0.269 0.146 1.000000e+00 1 ## Prox1os 2.605602e-05 1.6557742 0.053 0.013 1.000000e+00 1 ## Zfp217 2.625315e-05 0.1520923 0.388 0.238 1.000000e+00 1 ## Jrk 2.639506e-05 1.1626311 0.073 0.023 1.000000e+00 1 ## Snapin 2.660773e-05 0.5428909 0.310 0.186 1.000000e+00 1 ## Oas2 2.699203e-05 0.2620297 0.127 0.052 1.000000e+00 1 ## Ndufa61 2.702344e-05 0.1522351 0.980 0.787 1.000000e+00 1 ## Ebag9 2.714638e-05 0.2218105 0.412 0.254 1.000000e+00 1 ## Rtkn 2.721580e-05 0.1485969 0.388 0.235 1.000000e+00 1 ## Trappc13 2.737927e-05 0.2722593 0.498 0.320 1.000000e+00 1 ## Apoo 2.743160e-05 0.2859407 0.457 0.292 1.000000e+00 1 ## Caap1 2.751966e-05 0.3128411 0.298 0.170 1.000000e+00 1 ## Sdk1 2.772822e-05 2.1731569 0.045 0.010 1.000000e+00 1 ## Arxes2 2.782181e-05 1.2579277 0.122 0.051 1.000000e+00 1 ## Pcyox11 2.799011e-05 0.3201365 0.465 0.303 1.000000e+00 1 ## Sele 2.820364e-05 3.7926742 0.012 0.000 1.000000e+00 1 ## Gm5149 2.820364e-05 3.5324416 0.012 0.000 1.000000e+00 1 ## Cntfr 2.820364e-05 3.5214343 0.012 0.000 1.000000e+00 1 ## Frmpd1 2.820364e-05 3.7772641 0.012 0.000 1.000000e+00 1 ## Gm35855 2.820364e-05 3.7922469 0.012 0.000 1.000000e+00 1 ## Dtx1 2.820364e-05 3.7139665 0.012 0.000 1.000000e+00 1 ## Gm53330 2.820364e-05 3.9027321 0.012 0.000 1.000000e+00 1 ## Gm36210 2.820364e-05 3.5180470 0.012 0.000 1.000000e+00 1 ## Nat1 2.820364e-05 3.4864795 0.012 0.000 1.000000e+00 1 ## Gm27188 2.820364e-05 3.4943488 0.012 0.000 1.000000e+00 1 ## Gm10635 2.820364e-05 3.8585886 0.012 0.000 1.000000e+00 1 ## Rmst 2.820364e-05 3.6729820 0.012 0.000 1.000000e+00 1 ## Kcnip1 2.820364e-05 3.6939655 0.012 0.000 1.000000e+00 1 ## Rnf182 2.820364e-05 3.6634500 0.012 0.000 1.000000e+00 1 ## Gm34758 2.820364e-05 3.8893616 0.012 0.000 1.000000e+00 1 ## Gm41317 2.820364e-05 3.5161668 0.012 0.000 1.000000e+00 1 ## Gm53956 2.820364e-05 3.6480483 0.012 0.000 1.000000e+00 1 ## Gm53989 2.820364e-05 3.7109861 0.012 0.000 1.000000e+00 1 ## Gm47898 2.820364e-05 3.4995819 0.012 0.000 1.000000e+00 1 ## Entpd1 2.820364e-05 3.6065700 0.012 0.000 1.000000e+00 1 ## Gm32028 2.820364e-05 3.6819340 0.012 0.000 1.000000e+00 1 ## Il1rapl2 2.820364e-05 3.5735812 0.012 0.000 1.000000e+00 1 ## Gm15247 2.820364e-05 3.7385344 0.012 0.000 1.000000e+00 1 ## Rbm7 2.823470e-05 0.4089691 0.547 0.366 1.000000e+00 1 ## Clec11a 2.825362e-05 1.3353000 0.102 0.040 1.000000e+00 1 ## Cep41 2.856306e-05 1.0377877 0.139 0.063 1.000000e+00 1 ## Sumo11 2.885038e-05 0.1899177 0.906 0.687 1.000000e+00 1 ## Fbxo7 2.907531e-05 0.2740594 0.310 0.177 1.000000e+00 1 ## Zfp61 2.977626e-05 0.6420829 0.147 0.066 1.000000e+00 1 ## Tmem2191 2.983438e-05 0.3555750 0.576 0.383 1.000000e+00 1 ## Rab17 3.004227e-05 0.1798703 0.539 0.341 1.000000e+00 1 ## Emc2 3.006040e-05 0.2902857 0.527 0.340 1.000000e+00 1 ## Agap2 3.011092e-05 0.7737976 0.082 0.027 1.000000e+00 1 ## Stx2 3.025309e-05 0.7307476 0.143 0.064 1.000000e+00 1 ## Zfp72 3.035227e-05 1.7498552 0.057 0.015 1.000000e+00 1 ## Shroom2 3.037974e-05 0.6195903 0.188 0.097 1.000000e+00 1 ## Gm32440 3.054034e-05 0.9845140 0.073 0.023 1.000000e+00 1 ## Socs6 3.097555e-05 0.1231016 0.465 0.292 1.000000e+00 1 ## Usp101 3.111908e-05 0.2018928 0.461 0.288 1.000000e+00 1 ## Unc501 3.122813e-05 0.1612257 0.755 0.531 1.000000e+00 1 ## Vcf2 3.127736e-05 0.3454726 0.327 0.193 1.000000e+00 1 ## Sh3bp41 3.155958e-05 0.3092315 0.408 0.251 1.000000e+00 1 ## Acot6 3.158804e-05 1.6764329 0.057 0.015 1.000000e+00 1 ## Zfp791 3.162668e-05 1.5638435 0.045 0.010 1.000000e+00 1 ## Kdf1 3.174700e-05 0.2977123 0.384 0.230 1.000000e+00 1 ## Ubxn7 3.205963e-05 0.1099830 0.478 0.298 1.000000e+00 1 ## Zfp189 3.210182e-05 1.2167129 0.098 0.037 1.000000e+00 1 ## 5031425F14Rik 3.286563e-05 1.3572314 0.086 0.030 1.000000e+00 1 ## Ermard 3.287445e-05 0.2622837 0.245 0.133 1.000000e+00 1 ## Acad8 3.316590e-05 0.4605767 0.278 0.160 1.000000e+00 1 ## Gpatch111 3.340216e-05 0.1028879 0.551 0.356 1.000000e+00 1 ## Zkscan11 3.341936e-05 0.1563535 0.612 0.410 1.000000e+00 1 ## Gm154111 3.356907e-05 0.5022576 0.216 0.113 1.000000e+00 1 ## Insig2 3.362918e-05 0.1464351 0.555 0.375 1.000000e+00 1 ## Prkra 3.369396e-05 0.2342485 0.400 0.249 1.000000e+00 1 ## Slain2 3.384873e-05 0.1591232 0.588 0.405 1.000000e+00 1 ## Tgm7 3.391746e-05 3.4535236 0.016 0.001 1.000000e+00 1 ## Khdrbs2 3.414204e-05 3.3174258 0.016 0.001 1.000000e+00 1 ## Gm38985 3.414204e-05 3.7778188 0.016 0.001 1.000000e+00 1 ## Apoh 3.415559e-05 1.8101756 0.065 0.020 1.000000e+00 1 ## Gm10447 3.436803e-05 3.4592595 0.016 0.001 1.000000e+00 1 ## Tspan33 3.438693e-05 2.2889377 0.065 0.020 1.000000e+00 1 ## Eprs1 3.451616e-05 0.1304259 0.808 0.556 1.000000e+00 1 ## 4930447K03Rik 3.459545e-05 3.2751777 0.016 0.001 1.000000e+00 1 ## Cyp1b1 3.459545e-05 3.3378529 0.016 0.001 1.000000e+00 1 ## D6Ertd527e 3.482429e-05 3.1141529 0.016 0.001 1.000000e+00 1 ## Nwd1 3.482429e-05 2.3944357 0.016 0.001 1.000000e+00 1 ## Gm52101 3.482429e-05 2.7961883 0.016 0.001 1.000000e+00 1 ## 4833422M21Rik 3.482429e-05 3.0018609 0.016 0.001 1.000000e+00 1 ## Gm36587 3.482429e-05 3.3282012 0.016 0.001 1.000000e+00 1 ## Ocln1 3.483223e-05 0.1606925 0.755 0.542 1.000000e+00 1 ## Snhg17 3.483387e-05 0.5097442 0.212 0.110 1.000000e+00 1 ## Pard6a 3.490182e-05 0.7196975 0.180 0.089 1.000000e+00 1 ## Tmt1a 3.504796e-05 0.1198364 0.294 0.171 1.000000e+00 1 ## Car11 3.514376e-05 0.8456118 0.127 0.054 1.000000e+00 1 ## Zfp13 3.523240e-05 1.0905402 0.118 0.050 1.000000e+00 1 ## AU022252 3.529702e-05 0.4945410 0.245 0.136 1.000000e+00 1 ## Rec8 3.562769e-05 0.7795129 0.212 0.115 1.000000e+00 1 ## Msh2 3.571827e-05 0.3889889 0.273 0.153 1.000000e+00 1 ## Mllt3 3.585698e-05 0.2017801 0.449 0.298 1.000000e+00 1 ## Ptk2b 3.588728e-05 0.1949311 0.445 0.280 1.000000e+00 1 ## Zfp429 3.632212e-05 0.9926323 0.118 0.049 1.000000e+00 1 ## Polr1g 3.644725e-05 0.3402161 0.204 0.106 1.000000e+00 1 ## Snu13 3.645210e-05 0.1062814 0.767 0.529 1.000000e+00 1 ## Slc22a5 3.662729e-05 0.6711564 0.155 0.073 1.000000e+00 1 ## Eif2ak2 3.674342e-05 0.1528118 0.624 0.425 1.000000e+00 1 ## Eif2s11 3.812516e-05 0.1896543 0.669 0.447 1.000000e+00 1 ## Prr12 3.841353e-05 0.1244137 0.567 0.363 1.000000e+00 1 ## Ap3m1 3.851207e-05 0.1381627 0.498 0.330 1.000000e+00 1 ## Atp6v1e11 3.927880e-05 0.1620095 0.951 0.714 1.000000e+00 1 ## Hpcal11 3.931607e-05 0.1098824 0.718 0.493 1.000000e+00 1 ## Gm3740 3.966695e-05 1.6160576 0.049 0.012 1.000000e+00 1 ## Zmym3 3.975924e-05 0.2992490 0.286 0.161 1.000000e+00 1 ## Pdgfrl 3.977573e-05 1.9598013 0.049 0.012 1.000000e+00 1 ## Gm52313 3.984859e-05 1.0629338 0.078 0.026 1.000000e+00 1 ## Arsa 4.004543e-05 0.2089953 0.269 0.151 1.000000e+00 1 ## Pierce2 4.010525e-05 0.1893762 0.216 0.115 1.000000e+00 1 ## Btbd6 4.011333e-05 0.2733243 0.331 0.194 1.000000e+00 1 ## Hdac3 4.036607e-05 0.2306588 0.473 0.295 1.000000e+00 1 ## Coasy 4.053364e-05 0.2873401 0.363 0.219 1.000000e+00 1 ## Fbxo281 4.136872e-05 0.2124936 0.412 0.256 1.000000e+00 1 ## Ext2 4.150034e-05 0.4055617 0.351 0.215 1.000000e+00 1 ## Gm14399 4.152730e-05 0.9497747 0.102 0.040 1.000000e+00 1 ## Gm17224 4.155518e-05 1.8730350 0.049 0.012 1.000000e+00 1 ## Pigx 4.160388e-05 0.4119030 0.384 0.235 1.000000e+00 1 ## Tox41 4.174045e-05 0.2489910 0.571 0.385 1.000000e+00 1 ## Kpna3 4.199673e-05 0.1681065 0.433 0.278 1.000000e+00 1 ## Bod1 4.238553e-05 0.3908775 0.457 0.304 1.000000e+00 1 ## Znrf1 4.240906e-05 0.1313202 0.416 0.263 1.000000e+00 1 ## Tmem50b1 4.243267e-05 0.1103663 0.706 0.468 1.000000e+00 1 ## Phf20 4.245185e-05 0.1679170 0.461 0.290 1.000000e+00 1 ## Maea1 4.246332e-05 0.1388664 0.686 0.479 1.000000e+00 1 ## Dusp31 4.254590e-05 0.1708382 0.437 0.273 1.000000e+00 1 ## Rgn 4.256123e-05 1.9618632 0.082 0.029 1.000000e+00 1 ## Gpr52 4.286139e-05 0.3994043 0.061 0.017 1.000000e+00 1 ## Gspt2 4.310614e-05 0.4287013 0.192 0.098 1.000000e+00 1 ## Eid2b 4.339943e-05 0.5979761 0.216 0.115 1.000000e+00 1 ## Ahsa1 4.382363e-05 0.1245326 0.698 0.479 1.000000e+00 1 ## Zfp414 4.417152e-05 0.5818786 0.224 0.123 1.000000e+00 1 ## Smim18 4.428575e-05 1.6012762 0.041 0.008 1.000000e+00 1 ## Amer1 4.432326e-05 0.4434581 0.200 0.105 1.000000e+00 1 ## Gm31775 4.443466e-05 2.2374007 0.053 0.014 1.000000e+00 1 ## Smim12 4.478891e-05 0.1957253 0.261 0.152 1.000000e+00 1 ## Rinl 4.509484e-05 1.1070289 0.090 0.033 1.000000e+00 1 ## Rps19bp11 4.510786e-05 0.3150730 0.527 0.360 1.000000e+00 1 ## Gm42027 4.518257e-05 0.9269953 0.069 0.022 1.000000e+00 1 ## Zfp605 4.575039e-05 0.3862880 0.208 0.107 1.000000e+00 1 ## Ift81 4.581804e-05 0.2297761 0.363 0.216 1.000000e+00 1 ## Uqcc4 4.585561e-05 0.2586466 0.453 0.275 1.000000e+00 1 ## Gm53355 4.586874e-05 3.2032986 0.024 0.003 1.000000e+00 1 ## Creld1 4.590946e-05 0.4214765 0.192 0.098 1.000000e+00 1 ## Nalcn 4.605174e-05 1.8839779 0.053 0.014 1.000000e+00 1 ## Copb11 4.612276e-05 0.1577701 0.861 0.632 1.000000e+00 1 ## Rfesd 4.615264e-05 0.1542933 0.363 0.219 1.000000e+00 1 ## Zxdb 4.639321e-05 0.2688433 0.282 0.164 1.000000e+00 1 ## Slc12a1 4.650632e-05 3.0266783 0.024 0.003 1.000000e+00 1 ## Iqcb1 4.676197e-05 0.6226267 0.192 0.099 1.000000e+00 1 ## Cyp4f13 4.690328e-05 0.1401471 0.461 0.296 1.000000e+00 1 ## D930003E18Rik 4.693603e-05 2.9453588 0.024 0.003 1.000000e+00 1 ## 4930405A21Rik 4.715229e-05 2.6566977 0.024 0.003 1.000000e+00 1 ## Vnn3 4.715229e-05 2.7451971 0.024 0.003 1.000000e+00 1 ## Phactr3 4.736950e-05 2.6659925 0.024 0.003 1.000000e+00 1 ## Ldlr 4.765086e-05 0.1084706 0.661 0.457 1.000000e+00 1 ## D330022H12Rik 4.771094e-05 0.1352599 0.155 0.072 1.000000e+00 1 ## Tmc3 4.772451e-05 1.9599617 0.053 0.014 1.000000e+00 1 ## Unc93b1 4.868229e-05 0.1838433 0.396 0.246 1.000000e+00 1 ## Cstpp1 4.945320e-05 0.4071199 0.302 0.182 1.000000e+00 1 ## 2210016L21Rik1 4.965881e-05 0.1693100 0.743 0.539 1.000000e+00 1 ## Mrpl44 4.980318e-05 0.1653085 0.396 0.245 1.000000e+00 1 ## Gm31645 4.983321e-05 1.9097452 0.053 0.014 1.000000e+00 1 ## Elp51 4.994244e-05 0.2439804 0.453 0.284 1.000000e+00 1 ## Fra10ac1 5.048860e-05 0.2879625 0.392 0.247 1.000000e+00 1 ## Pdap11 5.070975e-05 0.1755977 0.976 0.804 1.000000e+00 1 ## Zfp369 5.087146e-05 0.2093361 0.269 0.152 1.000000e+00 1 ## C230037L18Rik 5.106655e-05 0.8571462 0.073 0.024 1.000000e+00 1 ## Nt5c2 5.115289e-05 0.1081604 0.429 0.266 1.000000e+00 1 ## Synj2 5.122111e-05 0.4612469 0.200 0.103 1.000000e+00 1 ## Degs21 5.122151e-05 0.1616389 0.959 0.761 1.000000e+00 1 ## Zfp1008 5.173298e-05 1.7997442 0.057 0.016 1.000000e+00 1 ## Zfp799 5.208869e-05 0.4275286 0.269 0.156 1.000000e+00 1 ## Xrcc51 5.231751e-05 0.2055045 0.376 0.226 1.000000e+00 1 ## Ube2a1 5.248960e-05 0.1731235 0.653 0.432 1.000000e+00 1 ## LTO1 5.263403e-05 0.2724755 0.339 0.208 1.000000e+00 1 ## Tmed1 5.265518e-05 1.1094334 0.155 0.077 1.000000e+00 1 ## Haus21 5.304669e-05 0.2770109 0.351 0.211 1.000000e+00 1 ## Srsf6 5.321427e-05 0.1213753 0.886 0.636 1.000000e+00 1 ## Gm30760 5.333271e-05 1.0195632 0.082 0.029 1.000000e+00 1 ## 4930526I15Rik 5.351525e-05 0.1286902 0.502 0.324 1.000000e+00 1 ## Gm39157 5.355631e-05 0.8132714 0.143 0.065 1.000000e+00 1 ## Cbr4 5.385418e-05 0.6046171 0.224 0.124 1.000000e+00 1 ## Igsf9b 5.388097e-05 0.4924341 0.143 0.067 1.000000e+00 1 ## Rpl10 5.531661e-05 0.1528507 1.000 0.904 1.000000e+00 1 ## Clrn31 5.547719e-05 0.1579749 0.804 0.576 1.000000e+00 1 ## Mapk9 5.550332e-05 0.4837517 0.282 0.166 1.000000e+00 1 ## Bpnt21 5.560859e-05 0.1000054 0.796 0.546 1.000000e+00 1 ## Prpf38a1 5.611277e-05 0.2364445 0.482 0.309 1.000000e+00 1 ## Sik2 5.618699e-05 0.2501799 0.208 0.112 1.000000e+00 1 ## Smim131 5.714741e-05 0.2005880 0.539 0.356 1.000000e+00 1 ## Tctn2 5.719686e-05 0.8259480 0.122 0.053 1.000000e+00 1 ## Nkapl 5.739166e-05 0.8533276 0.094 0.036 1.000000e+00 1 ## Qpctl 5.758942e-05 0.3039297 0.143 0.065 1.000000e+00 1 ## Ptprm 5.759212e-05 1.6265176 0.069 0.022 1.000000e+00 1 ## Terb1 5.771190e-05 0.9202875 0.053 0.014 1.000000e+00 1 ## Gm36446 5.815163e-05 2.1642060 0.033 0.006 1.000000e+00 1 ## Cfap298 5.848231e-05 0.2957301 0.351 0.216 1.000000e+00 1 ## Maf1 5.866125e-05 0.2161051 0.612 0.418 1.000000e+00 1 ## Chac2 5.872345e-05 0.2649872 0.282 0.168 1.000000e+00 1 ## Nfkbil1 5.888965e-05 0.2922421 0.351 0.222 1.000000e+00 1 ## Tmem125 5.907027e-05 0.9851809 0.106 0.043 1.000000e+00 1 ## Mmp16 5.924388e-05 0.8399262 0.106 0.043 1.000000e+00 1 ## Gm30603 5.942086e-05 2.3635401 0.033 0.006 1.000000e+00 1 ## Snx12 5.956954e-05 0.2365934 0.469 0.305 1.000000e+00 1 ## Gm37053 5.978924e-05 2.5270357 0.029 0.004 1.000000e+00 1 ## Surf2 5.986224e-05 0.2797601 0.298 0.172 1.000000e+00 1 ## Trit1 5.988167e-05 0.2762862 0.224 0.122 1.000000e+00 1 ## Gadd45gip11 6.000488e-05 0.1043872 0.735 0.502 1.000000e+00 1 ## Cpped1 6.009777e-05 0.1296996 0.494 0.316 1.000000e+00 1 ## Pmpcb 6.048035e-05 0.1651412 0.527 0.347 1.000000e+00 1 ## Gm54216 6.074952e-05 1.8753475 0.029 0.004 1.000000e+00 1 ## Myo1e1 6.091726e-05 0.1325135 0.567 0.367 1.000000e+00 1 ## Uqcc6 6.100771e-05 0.3468599 0.363 0.218 1.000000e+00 1 ## Ppp6c1 6.142021e-05 0.1737927 0.673 0.460 1.000000e+00 1 ## Gm34828 6.197042e-05 0.8750562 0.029 0.004 1.000000e+00 1 ## Tbc1d7 6.210191e-05 0.7547951 0.192 0.100 1.000000e+00 1 ## Gm46721 6.248516e-05 1.9703636 0.033 0.006 1.000000e+00 1 ## Rad9b 6.283449e-05 1.2411350 0.069 0.022 1.000000e+00 1 ## Lamtor11 6.312462e-05 0.2175692 0.776 0.567 1.000000e+00 1 ## Ly96 6.463523e-05 0.5898306 0.200 0.106 1.000000e+00 1 ## Nkiras1 6.480446e-05 0.1844053 0.482 0.313 1.000000e+00 1 ## Cog7 6.489093e-05 0.2133198 0.420 0.270 1.000000e+00 1 ## Mlkl 6.517255e-05 0.5273206 0.147 0.071 1.000000e+00 1 ## Bcdin3d 6.540210e-05 0.6192083 0.229 0.127 1.000000e+00 1 ## Pot1a 6.552614e-05 0.6018801 0.196 0.103 1.000000e+00 1 ## Smarcd11 6.558735e-05 0.1826458 0.445 0.291 1.000000e+00 1 ## Txnrd2 6.596130e-05 0.3361557 0.290 0.173 1.000000e+00 1 ## Vamp71 6.603530e-05 0.1726643 0.788 0.538 1.000000e+00 1 ## Blmh 6.712382e-05 0.2486847 0.531 0.373 1.000000e+00 1 ## Sh3bgrl2 6.743421e-05 0.8547405 0.131 0.061 1.000000e+00 1 ## Gm38944 6.765181e-05 1.5546658 0.045 0.010 1.000000e+00 1 ## Pxn1 6.791060e-05 0.2101148 0.702 0.475 1.000000e+00 1 ## Rce11 6.941580e-05 0.2068387 0.461 0.298 1.000000e+00 1 ## Magee1 6.959021e-05 0.6570665 0.110 0.045 1.000000e+00 1 ## Wdr13 6.982835e-05 0.1022763 0.612 0.423 1.000000e+00 1 ## Klhl42 7.046638e-05 0.1356405 0.351 0.213 1.000000e+00 1 ## 2210416O15Rik 7.112712e-05 1.4217572 0.065 0.020 1.000000e+00 1 ## Gm1976 7.117814e-05 0.8581950 0.094 0.036 1.000000e+00 1 ## C130013N14Rik 7.141947e-05 0.6982474 0.090 0.034 1.000000e+00 1 ## Cox15 7.246538e-05 0.3416365 0.208 0.111 1.000000e+00 1 ## Polm 7.252813e-05 0.1316428 0.290 0.167 1.000000e+00 1 ## Tmem170 7.308392e-05 0.2595075 0.322 0.196 1.000000e+00 1 ## Sesn3 7.338253e-05 0.2100488 0.220 0.117 1.000000e+00 1 ## Hexd 7.350372e-05 0.1868989 0.294 0.175 1.000000e+00 1 ## Gm13657 7.355236e-05 2.6636696 0.041 0.009 1.000000e+00 1 ## Rap2a 7.360175e-05 0.4596386 0.282 0.166 1.000000e+00 1 ## Zfp2601 7.501887e-05 0.1972322 0.543 0.369 1.000000e+00 1 ## Ddx60 7.513577e-05 0.1242205 0.314 0.182 1.000000e+00 1 ## Gm53283 7.564657e-05 0.5656133 0.127 0.057 1.000000e+00 1 ## Actr6 7.573493e-05 0.7356916 0.171 0.087 1.000000e+00 1 ## Carnmt11 7.597749e-05 0.1767434 0.486 0.320 1.000000e+00 1 ## Apmap1 7.597969e-05 0.3343319 0.404 0.253 1.000000e+00 1 ## Mta1 7.686186e-05 0.1592415 0.331 0.201 1.000000e+00 1 ## Fbxo25 7.738784e-05 0.1893744 0.686 0.477 1.000000e+00 1 ## Krba1 7.809511e-05 0.4719915 0.127 0.056 1.000000e+00 1 ## Ift122 7.847824e-05 0.2616375 0.245 0.134 1.000000e+00 1 ## Mmp23 7.943337e-05 0.2353547 0.176 0.087 1.000000e+00 1 ## Gm42149 8.110996e-05 2.2227229 0.041 0.009 1.000000e+00 1 ## Trappc11 8.147835e-05 0.1487527 0.482 0.310 1.000000e+00 1 ## Thumpd2 8.195636e-05 0.8812301 0.110 0.047 1.000000e+00 1 ## Lrrc27 8.329515e-05 1.5445646 0.041 0.009 1.000000e+00 1 ## Dnajb111 8.347152e-05 0.1888360 0.792 0.556 1.000000e+00 1 ## Zmym1 8.396963e-05 0.6073656 0.180 0.095 1.000000e+00 1 ## Pik3c2g 8.443395e-05 0.6439289 0.086 0.031 1.000000e+00 1 ## Snhg3 8.457660e-05 0.1511139 0.445 0.277 1.000000e+00 1 ## Trim37 8.510596e-05 0.2794160 0.302 0.179 1.000000e+00 1 ## Ogg1 8.529588e-05 0.2773499 0.114 0.050 1.000000e+00 1 ## Ndufs31 8.560741e-05 0.1166562 0.857 0.614 1.000000e+00 1 ## Cdo1 8.561034e-05 0.5266885 0.155 0.075 1.000000e+00 1 ## Ppp1r15a 8.633381e-05 0.1138477 0.449 0.292 1.000000e+00 1 ## P2ry61 8.746531e-05 0.9448545 0.086 0.032 1.000000e+00 1 ## Gabarap1 8.750418e-05 0.1221183 1.000 0.874 1.000000e+00 1 ## Gm51556 8.787997e-05 0.2281663 0.371 0.233 1.000000e+00 1 ## Zfp810 8.791427e-05 0.2948032 0.229 0.124 1.000000e+00 1 ## Usp11 8.793449e-05 0.2262721 0.163 0.079 1.000000e+00 1 ## 9130604C24Rik 8.795080e-05 1.1862862 0.082 0.029 1.000000e+00 1 ## Mknk2 8.828145e-05 0.1268794 0.490 0.313 1.000000e+00 1 ## Gm31793 8.848657e-05 1.8496072 0.057 0.017 1.000000e+00 1 ## Cnot9 8.959614e-05 0.3784851 0.229 0.128 1.000000e+00 1 ## Gdi21 8.967365e-05 0.1456132 0.959 0.737 1.000000e+00 1 ## Shc41 8.999749e-05 0.3949638 0.139 0.062 1.000000e+00 1 ## Tymp 9.018934e-05 1.5121720 0.041 0.009 1.000000e+00 1 ## Hace1 9.036249e-05 0.3137750 0.278 0.161 1.000000e+00 1 ## Hspa2 9.100894e-05 0.7653911 0.102 0.042 1.000000e+00 1 ## Mknk1 9.102450e-05 0.1840449 0.282 0.166 1.000000e+00 1 ## Psmb61 9.124410e-05 0.1001803 0.980 0.747 1.000000e+00 1 ## Ppip5k21 9.190618e-05 0.1221513 0.731 0.510 1.000000e+00 1 ## Brms1 9.512775e-05 0.1996779 0.482 0.317 1.000000e+00 1 ## Dclre1b 9.545871e-05 1.2366833 0.106 0.045 1.000000e+00 1 ## Zscan21 9.650462e-05 0.2922026 0.245 0.136 1.000000e+00 1 ## Uhmk11 9.675128e-05 0.1087197 0.682 0.469 1.000000e+00 1 ## Nif3l1 9.720194e-05 0.6547820 0.212 0.119 1.000000e+00 1 ## Bmp6 9.905447e-05 2.8190736 0.037 0.008 1.000000e+00 1 ## Prss36 9.946670e-05 0.6925957 0.135 0.062 1.000000e+00 1 ## Ddah1 9.952677e-05 0.8136091 0.106 0.045 1.000000e+00 1 ## Zfp775 1.001243e-04 0.5125405 0.118 0.052 1.000000e+00 1 ## Bmt2 1.002051e-04 0.1920440 0.437 0.271 1.000000e+00 1 ## Cbl 1.010229e-04 0.1152031 0.702 0.526 1.000000e+00 1 ## Lmf1 1.024113e-04 0.5729494 0.310 0.194 1.000000e+00 1 ## Coq6 1.024549e-04 0.1324224 0.310 0.188 1.000000e+00 1 ## Cdc731 1.027385e-04 0.2286042 0.718 0.517 1.000000e+00 1 ## Bax1 1.034230e-04 0.2369584 0.894 0.649 1.000000e+00 1 ## Ankrd28 1.035223e-04 0.3173924 0.282 0.168 1.000000e+00 1 ## Stau2 1.043286e-04 0.4304543 0.359 0.229 1.000000e+00 1 ## Zfp119a 1.056290e-04 0.5648811 0.143 0.067 1.000000e+00 1 ## Nphp3 1.093045e-04 1.0048490 0.094 0.038 1.000000e+00 1 ## Slc33a11 1.100236e-04 0.1318089 0.669 0.477 1.000000e+00 1 ## Prrc2b1 1.105917e-04 0.1503607 0.845 0.606 1.000000e+00 1 ## Dph5 1.117430e-04 0.6745923 0.176 0.092 1.000000e+00 1 ## Etaa1os 1.119982e-04 1.0001606 0.086 0.033 1.000000e+00 1 ## Lrp8os2 1.132785e-04 1.6249950 0.037 0.008 1.000000e+00 1 ## Sin3b1 1.132938e-04 0.1867771 0.780 0.547 1.000000e+00 1 ## Stip11 1.135220e-04 0.1700334 0.641 0.425 1.000000e+00 1 ## Ric8b 1.139822e-04 0.2094084 0.359 0.223 1.000000e+00 1 ## Snhg6 1.145773e-04 0.4327223 0.245 0.142 1.000000e+00 1 ## Zfp661 1.151318e-04 0.9948878 0.078 0.028 1.000000e+00 1 ## Snd1 1.156364e-04 0.2176631 0.751 0.544 1.000000e+00 1 ## 1110020A21Rik 1.158370e-04 1.3539811 0.098 0.041 1.000000e+00 1 ## Stambp 1.161413e-04 0.1205103 0.449 0.292 1.000000e+00 1 ## Plekha4 1.169880e-04 0.8746099 0.069 0.023 1.000000e+00 1 ## Krt23 1.179498e-04 0.6756736 0.073 0.026 1.000000e+00 1 ## Gm5431 1.194165e-04 1.7795717 0.037 0.008 1.000000e+00 1 ## Umps1 1.198013e-04 0.2131595 0.424 0.265 1.000000e+00 1 ## Dlc1 1.201463e-04 1.1025803 0.106 0.045 1.000000e+00 1 ## Actr3b 1.205338e-04 4.1096094 0.020 0.002 1.000000e+00 1 ## Zbtb1 1.208302e-04 0.1382529 0.412 0.263 1.000000e+00 1 ## Gm46410 1.211234e-04 3.1210814 0.020 0.002 1.000000e+00 1 ## Mrpl211 1.213547e-04 0.1281464 0.776 0.539 1.000000e+00 1 ## Psma61 1.214252e-04 0.1501284 0.898 0.632 1.000000e+00 1 ## Slc2a3 1.217157e-04 1.5264411 0.020 0.002 1.000000e+00 1 ## Sspn 1.217157e-04 3.0563826 0.020 0.002 1.000000e+00 1 ## Pfdn61 1.221954e-04 0.1173991 0.865 0.636 1.000000e+00 1 ## Gm33447 1.229086e-04 2.8686782 0.020 0.002 1.000000e+00 1 ## Ly6f 1.229086e-04 2.9320402 0.020 0.002 1.000000e+00 1 ## Cmpk2 1.236361e-04 0.2761062 0.220 0.123 1.000000e+00 1 ## Gm33130 1.241124e-04 3.2568148 0.020 0.002 1.000000e+00 1 ## Crls11 1.246662e-04 0.1310888 0.465 0.297 1.000000e+00 1 ## Nck11 1.253676e-04 0.1794702 0.522 0.349 1.000000e+00 1 ## Gm35601 1.259391e-04 2.1811777 0.020 0.002 1.000000e+00 1 ## Gm36790 1.259391e-04 0.3097131 0.020 0.002 1.000000e+00 1 ## Tmed21 1.266101e-04 0.1573781 0.943 0.733 1.000000e+00 1 ## Ap4b1 1.282465e-04 0.4654278 0.237 0.136 1.000000e+00 1 ## Dtx2 1.294810e-04 0.5536768 0.163 0.082 1.000000e+00 1 ## Cebpz 1.310448e-04 0.1647902 0.661 0.458 1.000000e+00 1 ## 6030458C11Rik 1.315051e-04 0.1949009 0.339 0.213 1.000000e+00 1 ## Mrps23 1.321840e-04 0.3299711 0.498 0.331 1.000000e+00 1 ## Ercc1 1.328026e-04 0.5596354 0.237 0.133 1.000000e+00 1 ## Ythdf11 1.336207e-04 0.1727550 0.584 0.400 1.000000e+00 1 ## Dhps 1.336360e-04 0.2389972 0.527 0.351 1.000000e+00 1 ## Mtfr1 1.372712e-04 0.2658897 0.359 0.234 1.000000e+00 1 ## Mthfd1 1.375200e-04 0.2332258 0.269 0.154 1.000000e+00 1 ## Otulin 1.382693e-04 0.1011292 0.682 0.480 1.000000e+00 1 ## G6pc3 1.428008e-04 0.1646727 0.490 0.311 1.000000e+00 1 ## Yae1d1 1.431946e-04 0.1132627 0.347 0.210 1.000000e+00 1 ## Mrpl2 1.439094e-04 0.1501356 0.539 0.338 1.000000e+00 1 ## Exosc1 1.445932e-04 0.1473540 0.318 0.191 1.000000e+00 1 ## Gm32998 1.447452e-04 2.3346248 0.041 0.010 1.000000e+00 1 ## Cox18 1.448102e-04 0.4834370 0.269 0.159 1.000000e+00 1 ## Capn10 1.448439e-04 0.4160458 0.282 0.171 1.000000e+00 1 ## Aida 1.449841e-04 0.2293142 0.376 0.239 1.000000e+00 1 ## Hhat 1.452992e-04 1.5178277 0.065 0.022 1.000000e+00 1 ## Ndufaf7 1.457216e-04 0.2769524 0.306 0.184 1.000000e+00 1 ## Gm39807 1.468295e-04 1.2037488 0.033 0.006 1.000000e+00 1 ## Wdr89 1.477408e-04 0.1005121 0.367 0.226 1.000000e+00 1 ## Paqr7 1.496687e-04 0.8082525 0.118 0.054 1.000000e+00 1 ## Zhx3 1.509617e-04 0.1466246 0.245 0.136 1.000000e+00 1 ## Bst2 1.517017e-04 0.5936600 0.159 0.082 1.000000e+00 1 ## B230217C12Rik 1.527992e-04 1.3032474 0.061 0.020 1.000000e+00 1 ## Pim2 1.528927e-04 0.1018855 0.376 0.229 1.000000e+00 1 ## Sinhcaf1 1.533189e-04 0.1090229 0.571 0.386 1.000000e+00 1 ## Rnf122 1.539296e-04 2.1844340 0.041 0.010 1.000000e+00 1 ## 4930481A15Rik 1.540910e-04 0.4205069 0.237 0.136 1.000000e+00 1 ## Ring1 1.552344e-04 0.2832535 0.343 0.212 1.000000e+00 1 ## Klhl12 1.561794e-04 0.3303938 0.265 0.157 1.000000e+00 1 ## Klf131 1.565986e-04 0.1342165 0.792 0.563 1.000000e+00 1 ## Sipa1 1.577266e-04 0.4684039 0.155 0.075 1.000000e+00 1 ## B230369F24Rik 1.585354e-04 0.5068719 0.110 0.048 1.000000e+00 1 ## Tor1aip11 1.646832e-04 0.1612224 0.563 0.384 1.000000e+00 1 ## Tmem170b 1.651371e-04 0.4089521 0.278 0.166 1.000000e+00 1 ## Gm38521 1.661630e-04 0.9186678 0.151 0.076 1.000000e+00 1 ## Gm53861 1.674358e-04 2.5752698 0.029 0.005 1.000000e+00 1 ## Cd320 1.679887e-04 0.3077355 0.404 0.265 1.000000e+00 1 ## Nsmce4a 1.695143e-04 0.1274740 0.612 0.416 1.000000e+00 1 ## Gm21145 1.698808e-04 2.2580404 0.029 0.005 1.000000e+00 1 ## Scn7a 1.711158e-04 2.3050886 0.029 0.005 1.000000e+00 1 ## Cyp2c38 1.717365e-04 2.5570077 0.029 0.005 1.000000e+00 1 ## Igfals 1.721727e-04 0.6513271 0.106 0.045 1.000000e+00 1 ## Cnpy3 1.724909e-04 0.3973272 0.322 0.200 1.000000e+00 1 ## Snx23 1.726824e-04 0.1066936 0.727 0.509 1.000000e+00 1 ## Klhl91 1.729344e-04 0.1715375 0.690 0.483 1.000000e+00 1 ## Gm92221 1.735521e-04 0.1433570 0.429 0.275 1.000000e+00 1 ## Fam110c 1.740914e-04 0.4033232 0.200 0.110 1.000000e+00 1 ## Zfp280b 1.745527e-04 0.2584747 0.367 0.232 1.000000e+00 1 ## Pgpep11 1.749453e-04 0.2545020 0.527 0.357 1.000000e+00 1 ## Rnf1031 1.757277e-04 0.1273411 0.780 0.567 1.000000e+00 1 ## Fam32a1 1.760041e-04 0.1119303 0.853 0.611 1.000000e+00 1 ## Rapgef1 1.764078e-04 0.1243054 0.306 0.184 1.000000e+00 1 ## Adgre5 1.767649e-04 0.5804426 0.192 0.105 1.000000e+00 1 ## Nme4 1.769093e-04 1.3909822 0.114 0.053 1.000000e+00 1 ## Gm13822 1.775301e-04 1.0587459 0.065 0.022 1.000000e+00 1 ## Cnnm3 1.793350e-04 0.2960726 0.273 0.165 1.000000e+00 1 ## Cd86 1.798551e-04 1.8892212 0.024 0.003 1.000000e+00 1 ## Hectd2os 1.800005e-04 2.3664394 0.029 0.005 1.000000e+00 1 ## Samd101 1.807687e-04 0.1757842 0.465 0.312 1.000000e+00 1 ## Fut4 1.809430e-04 1.0937530 0.122 0.058 1.000000e+00 1 ## Tbl2 1.817048e-04 0.4698153 0.265 0.159 1.000000e+00 1 ## Cln5 1.819818e-04 0.1341772 0.180 0.094 1.000000e+00 1 ## Gstp3 1.831148e-04 0.8459490 0.180 0.100 1.000000e+00 1 ## 5830448L01Rik 1.836363e-04 0.4864043 0.163 0.085 1.000000e+00 1 ## Pex11b 1.845082e-04 0.5190014 0.261 0.156 1.000000e+00 1 ## Slc9a61 1.852566e-04 0.3826576 0.237 0.136 1.000000e+00 1 ## Mvd 1.873200e-04 0.5484921 0.241 0.143 1.000000e+00 1 ## Ccdc181 1.880102e-04 0.2711042 0.302 0.183 1.000000e+00 1 ## Zfp874a 1.883138e-04 0.4855819 0.139 0.066 1.000000e+00 1 ## Gm14295 1.884822e-04 0.9911608 0.127 0.060 1.000000e+00 1 ## Btc 1.904271e-04 0.3207408 0.229 0.133 1.000000e+00 1 ## Fkbp81 1.915550e-04 0.1192660 0.967 0.730 1.000000e+00 1 ## Eef1akmt4 1.927795e-04 0.9021796 0.114 0.052 1.000000e+00 1 ## Zfp511 1.951511e-04 0.6906466 0.282 0.171 1.000000e+00 1 ## Golt1b 1.996838e-04 0.4569347 0.294 0.182 1.000000e+00 1 ## Rras21 2.004217e-04 0.1833121 0.424 0.283 1.000000e+00 1 ## Tpd52l2 2.006174e-04 0.1072527 0.527 0.354 1.000000e+00 1 ## Npb 2.021778e-04 1.7085552 0.045 0.012 1.000000e+00 1 ## Mrnip 2.062639e-04 1.3411544 0.086 0.034 1.000000e+00 1 ## Poc1b1 2.083962e-04 0.1891450 0.522 0.358 1.000000e+00 1 ## Dhrs13 2.114956e-04 0.8287585 0.114 0.052 1.000000e+00 1 ## Fbxl161 2.133899e-04 0.1639254 0.457 0.296 1.000000e+00 1 ## Manbal1 2.152970e-04 0.1960887 0.449 0.300 1.000000e+00 1 ## Kics2 2.170088e-04 0.5173850 0.073 0.027 1.000000e+00 1 ## Ppp1r351 2.173094e-04 0.2726541 0.437 0.296 1.000000e+00 1 ## Wdr19 2.173667e-04 0.6330862 0.098 0.041 1.000000e+00 1 ## Gimap11 2.187210e-04 0.2234124 0.620 0.453 1.000000e+00 1 ## Gm43653 2.187379e-04 1.0853387 0.045 0.012 1.000000e+00 1 ## Toe1 2.211385e-04 0.3830490 0.265 0.158 1.000000e+00 1 ## 2610528J11Rik1 2.216026e-04 0.2147998 0.645 0.446 1.000000e+00 1 ## Acot2 2.227647e-04 0.5972114 0.163 0.086 1.000000e+00 1 ## Pgam5 2.228963e-04 0.3044459 0.278 0.170 1.000000e+00 1 ## Exoc8 2.229082e-04 0.1727405 0.363 0.238 1.000000e+00 1 ## Mpzl1 2.229534e-04 0.2063797 0.514 0.339 1.000000e+00 1 ## Tgfa 2.238330e-04 0.6173812 0.188 0.103 1.000000e+00 1 ## Gm10190 2.238415e-04 1.2395546 0.049 0.014 1.000000e+00 1 ## Ydjc 2.241053e-04 0.2936411 0.327 0.206 1.000000e+00 1 ## Gm9776 2.250200e-04 1.2901822 0.078 0.029 1.000000e+00 1 ## Pdcd11 2.255880e-04 0.1164819 0.522 0.347 1.000000e+00 1 ## Mcph1 2.262675e-04 0.2517347 0.224 0.129 1.000000e+00 1 ## Morn4 2.267293e-04 0.5228321 0.106 0.045 1.000000e+00 1 ## Lmf2 2.270630e-04 0.3401319 0.261 0.157 1.000000e+00 1 ## Ttll12 2.279889e-04 0.7887782 0.200 0.112 1.000000e+00 1 ## Pspc1 2.292374e-04 0.1099805 0.343 0.216 1.000000e+00 1 ## C2cd4d 2.297829e-04 2.1444660 0.037 0.008 1.000000e+00 1 ## Rnf139 2.300680e-04 0.2315835 0.310 0.189 1.000000e+00 1 ## Gm346861 2.307560e-04 0.6050217 0.114 0.051 1.000000e+00 1 ## Gpatch2l 2.312540e-04 0.2130786 0.261 0.157 1.000000e+00 1 ## Cep97 2.332524e-04 0.3694568 0.155 0.078 1.000000e+00 1 ## Sike11 2.334949e-04 0.2045048 0.457 0.308 1.000000e+00 1 ## Ccdc127 2.343799e-04 0.2467239 0.494 0.336 1.000000e+00 1 ## Tgif1 2.352294e-04 0.1974451 0.257 0.153 1.000000e+00 1 ## Dnajc25 2.358284e-04 0.1388216 0.376 0.240 1.000000e+00 1 ## Lancl2 2.358416e-04 0.6771990 0.204 0.117 1.000000e+00 1 ## Trim67 2.376567e-04 1.1583110 0.061 0.020 1.000000e+00 1 ## Ncdn 2.404594e-04 0.1011020 0.192 0.104 1.000000e+00 1 ## Arrdc3 2.413298e-04 0.1912550 0.327 0.205 1.000000e+00 1 ## Crbn1 2.429028e-04 0.2020465 0.441 0.293 1.000000e+00 1 ## Fbxw21 2.438926e-04 0.1756406 0.531 0.359 1.000000e+00 1 ## Rad171 2.441002e-04 0.1100596 0.494 0.322 1.000000e+00 1 ## Tmco11 2.444010e-04 0.1028901 0.894 0.637 1.000000e+00 1 ## Nap1l3 2.445029e-04 0.5622371 0.139 0.068 1.000000e+00 1 ## Plin21 2.446026e-04 0.1465615 0.600 0.403 1.000000e+00 1 ## Zfp780b 2.450905e-04 0.1936258 0.302 0.181 1.000000e+00 1 ## F8 2.464987e-04 0.7938321 0.090 0.036 1.000000e+00 1 ## Man2c11 2.481288e-04 0.1283980 0.441 0.292 1.000000e+00 1 ## Polr3g 2.503850e-04 0.8325011 0.159 0.084 1.000000e+00 1 ## Tgds 2.506683e-04 0.6347859 0.224 0.130 1.000000e+00 1 ## Zbtb8os 2.513893e-04 0.2161497 0.543 0.370 1.000000e+00 1 ## Pskh1 2.538667e-04 0.3951954 0.282 0.177 1.000000e+00 1 ## Gm31728 2.543320e-04 1.3869747 0.086 0.035 1.000000e+00 1 ## Gm15489 2.557569e-04 0.3878603 0.224 0.130 1.000000e+00 1 ## Carm1 2.579468e-04 0.2603672 0.318 0.198 1.000000e+00 1 ## Rab101 2.587654e-04 0.1141179 0.898 0.707 1.000000e+00 1 ## Rpl7l11 2.603104e-04 0.1941569 0.527 0.347 1.000000e+00 1 ## Gm52134 2.610449e-04 0.5483427 0.037 0.008 1.000000e+00 1 ## Esam 2.611039e-04 0.4229993 0.151 0.077 1.000000e+00 1 ## Arpc1b1 2.614838e-04 0.1420905 0.959 0.789 1.000000e+00 1 ## Agt 2.622988e-04 0.8290086 0.078 0.029 1.000000e+00 1 ## Agps 2.633772e-04 0.1247853 0.335 0.212 1.000000e+00 1 ## 0610009L18Rik 2.637263e-04 0.6938938 0.155 0.081 1.000000e+00 1 ## Ttc5 2.650098e-04 0.1669358 0.469 0.305 1.000000e+00 1 ## E2f6 2.652254e-04 0.3662276 0.233 0.134 1.000000e+00 1 ## Prelid2 2.664879e-04 0.4332891 0.286 0.180 1.000000e+00 1 ## Utp61 2.688587e-04 0.4012330 0.290 0.176 1.000000e+00 1 ## 1810021B22Rik 2.699829e-04 1.1988106 0.082 0.032 1.000000e+00 1 ## Mypop 2.710860e-04 0.7187911 0.131 0.063 1.000000e+00 1 ## Pdcd2l 2.714938e-04 0.4991685 0.278 0.171 1.000000e+00 1 ## Map1lc3b1 2.749511e-04 0.1110236 0.902 0.712 1.000000e+00 1 ## Gm31619 2.752415e-04 0.7405179 0.061 0.020 1.000000e+00 1 ## Rab11bos1 2.755981e-04 0.3608338 0.216 0.124 1.000000e+00 1 ## Atn1 2.760327e-04 0.1881463 0.510 0.349 1.000000e+00 1 ## Gm39499 2.771544e-04 0.5383714 0.053 0.016 1.000000e+00 1 ## Jmjd4 2.775343e-04 1.2750062 0.135 0.068 1.000000e+00 1 ## Tmem70 2.802245e-04 0.2851108 0.453 0.303 1.000000e+00 1 ## 1110025M09Rik 2.824953e-04 0.8891582 0.086 0.034 1.000000e+00 1 ## Csad 2.855476e-04 0.1762110 0.433 0.279 1.000000e+00 1 ## 5830433I10Rik 2.861594e-04 0.1172571 0.167 0.086 1.000000e+00 1 ## Dqx1 2.937608e-04 0.1048554 0.371 0.236 1.000000e+00 1 ## Gm43568 2.940401e-04 1.9971727 0.041 0.010 1.000000e+00 1 ## Commd2 2.962915e-04 0.1029143 0.469 0.296 1.000000e+00 1 ## Washc3 2.971008e-04 0.3094688 0.490 0.338 1.000000e+00 1 ## Erfl 2.989603e-04 3.4404761 0.016 0.001 1.000000e+00 1 ## Tomm40l 3.017369e-04 0.1355591 0.290 0.175 1.000000e+00 1 ## Pabpn1l 3.021645e-04 2.8471713 0.016 0.001 1.000000e+00 1 ## Ube2d31 3.034857e-04 0.1285058 0.992 0.809 1.000000e+00 1 ## Zfp60 3.037650e-04 0.3772218 0.224 0.129 1.000000e+00 1 ## Ndn 3.037787e-04 3.0476324 0.016 0.001 1.000000e+00 1 ## 1700020N18Rik 3.054009e-04 3.0671884 0.016 0.001 1.000000e+00 1 ## Gm50595 3.054009e-04 0.7561792 0.016 0.001 1.000000e+00 1 ## Ccdc89 3.054009e-04 2.8752382 0.016 0.001 1.000000e+00 1 ## Gm51715 3.070313e-04 2.9673897 0.016 0.001 1.000000e+00 1 ## Akr1c18 3.070313e-04 2.1343078 0.016 0.001 1.000000e+00 1 ## Trim17 3.071299e-04 0.5038526 0.261 0.155 1.000000e+00 1 ## Gm31305 3.086698e-04 1.6157327 0.016 0.001 1.000000e+00 1 ## Gm52781 3.086698e-04 2.8513928 0.016 0.001 1.000000e+00 1 ## Gm36223 3.086698e-04 2.7543248 0.016 0.001 1.000000e+00 1 ## Adamts9 3.086698e-04 1.9669670 0.016 0.001 1.000000e+00 1 ## Gm51458 3.086698e-04 2.7560829 0.016 0.001 1.000000e+00 1 ## F420014N23Rik 3.086698e-04 2.3103270 0.016 0.001 1.000000e+00 1 ## Selenoo 3.094823e-04 0.3522312 0.261 0.159 1.000000e+00 1 ## Xpo4 3.130309e-04 0.2095157 0.339 0.208 1.000000e+00 1 ## Atf6b1 3.138680e-04 0.1961239 0.412 0.268 1.000000e+00 1 ## Rasa11 3.140706e-04 0.1947447 0.706 0.510 1.000000e+00 1 ## Zfp78 3.146401e-04 1.3091229 0.065 0.023 1.000000e+00 1 ## Slc25a38 3.155707e-04 0.4029541 0.237 0.142 1.000000e+00 1 ## Kcnh7 3.159290e-04 0.7800411 0.102 0.045 1.000000e+00 1 ## Tsen2 3.190885e-04 1.2066588 0.114 0.054 1.000000e+00 1 ## Zbtb32 3.197853e-04 0.8987415 0.041 0.010 1.000000e+00 1 ## Gm420671 3.229382e-04 0.6770510 0.167 0.089 1.000000e+00 1 ## Pcdhb5 3.235317e-04 1.9815396 0.033 0.007 1.000000e+00 1 ## Rmnd5b 3.260049e-04 0.4979897 0.261 0.157 1.000000e+00 1 ## Nsmf 3.262500e-04 0.1152831 0.384 0.240 1.000000e+00 1 ## Ebi3 3.306469e-04 1.0454733 0.078 0.030 1.000000e+00 1 ## Zc3h6 3.312443e-04 0.3176982 0.171 0.091 1.000000e+00 1 ## Osbpl6 3.312590e-04 0.1791577 0.314 0.195 1.000000e+00 1 ## Asnsd1 3.321012e-04 0.1848159 0.355 0.222 1.000000e+00 1 ## Tbc1d19 3.328729e-04 0.2516723 0.143 0.072 1.000000e+00 1 ## Gm42253 3.334353e-04 1.5187277 0.041 0.010 1.000000e+00 1 ## Cep78 3.350316e-04 0.4123450 0.216 0.126 1.000000e+00 1 ## Kdm4a 3.359138e-04 0.1109448 0.400 0.261 1.000000e+00 1 ## Thnsl1 3.369076e-04 0.3936809 0.176 0.093 1.000000e+00 1 ## Cldn61 3.410218e-04 0.5510817 0.176 0.094 1.000000e+00 1 ## Appbp21 3.411189e-04 0.2278399 0.547 0.374 1.000000e+00 1 ## Eola1 3.421635e-04 0.3535575 0.335 0.216 1.000000e+00 1 ## Gm41231 3.422414e-04 1.3424159 0.041 0.010 1.000000e+00 1 ## 5830454E08Rik 3.430286e-04 0.8863390 0.114 0.053 1.000000e+00 1 ## Uck2 3.449687e-04 0.5408589 0.171 0.094 1.000000e+00 1 ## Ttl 3.486017e-04 0.7096728 0.122 0.059 1.000000e+00 1 ## B3gnt8 3.501861e-04 1.3056285 0.061 0.021 1.000000e+00 1 ## Cfap300 3.516965e-04 1.7939053 0.053 0.017 1.000000e+00 1 ## Rpgrip1l 3.526531e-04 0.3916884 0.208 0.119 1.000000e+00 1 ## Tefm 3.529625e-04 0.6634514 0.192 0.108 1.000000e+00 1 ## Gm32013 3.537098e-04 1.1900133 0.045 0.013 1.000000e+00 1 ## Polr3gl 3.559272e-04 0.1215155 0.371 0.238 1.000000e+00 1 ## Usp32 3.563775e-04 0.1035690 0.437 0.285 1.000000e+00 1 ## Pno1 3.578385e-04 0.3048747 0.376 0.238 1.000000e+00 1 ## Zfp111 3.617288e-04 0.4949757 0.216 0.125 1.000000e+00 1 ## Sfxn3 3.665928e-04 0.3030551 0.163 0.086 1.000000e+00 1 ## Pcsk2os1 3.692113e-04 0.6605961 0.122 0.058 1.000000e+00 1 ## AA388235 3.702754e-04 1.3849178 0.057 0.019 1.000000e+00 1 ## Gna12 3.715288e-04 1.5635827 0.049 0.015 1.000000e+00 1 ## Stx81 3.724799e-04 0.1637095 0.592 0.425 1.000000e+00 1 ## Ubl4a 3.769807e-04 0.3604218 0.298 0.187 1.000000e+00 1 ## Gm30906 3.783274e-04 0.5671832 0.082 0.032 1.000000e+00 1 ## Trim321 3.794960e-04 0.2479131 0.233 0.132 1.000000e+00 1 ## Bscl2 3.806159e-04 0.1208682 0.453 0.304 1.000000e+00 1 ## Tmem17 3.807468e-04 1.3380392 0.069 0.026 1.000000e+00 1 ## Ccdc159 3.843273e-04 1.3667695 0.049 0.015 1.000000e+00 1 ## Samd5 3.845152e-04 0.3394862 0.069 0.025 1.000000e+00 1 ## Abhd10 3.869825e-04 0.1597013 0.310 0.194 1.000000e+00 1 ## Ksr2 3.894811e-04 1.1095827 0.045 0.013 1.000000e+00 1 ## Rbbp7 3.929873e-04 0.1252706 0.902 0.669 1.000000e+00 1 ## Nup62 3.941954e-04 0.3119820 0.245 0.143 1.000000e+00 1 ## Usp31 3.942376e-04 0.1787527 0.204 0.116 1.000000e+00 1 ## Pold3 3.967430e-04 0.1270046 0.371 0.235 1.000000e+00 1 ## Vsig10l 3.972410e-04 1.1971022 0.053 0.017 1.000000e+00 1 ## Nup133 4.007585e-04 0.9245709 0.147 0.078 1.000000e+00 1 ## Nosip 4.023097e-04 0.2820809 0.351 0.231 1.000000e+00 1 ## 1700123O20Rik 4.032664e-04 0.2293513 0.473 0.317 1.000000e+00 1 ## Nudt1 4.038646e-04 0.4305999 0.253 0.151 1.000000e+00 1 ## Gm39342 4.053319e-04 0.5110630 0.220 0.130 1.000000e+00 1 ## Slc31a21 4.073572e-04 0.2816180 0.269 0.161 1.000000e+00 1 ## Chchd5 4.109787e-04 0.2753941 0.314 0.198 1.000000e+00 1 ## Zfp2661 4.117617e-04 0.1554889 0.535 0.370 1.000000e+00 1 ## Pigu1 4.142869e-04 0.2474313 0.388 0.258 1.000000e+00 1 ## Eml2 4.146759e-04 0.2897011 0.322 0.204 1.000000e+00 1 ## Trim141 4.180431e-04 0.1421516 0.457 0.312 1.000000e+00 1 ## 9430065F17Rik 4.278980e-04 0.7838779 0.082 0.033 1.000000e+00 1 ## Rpusd4 4.295325e-04 0.7283130 0.176 0.096 1.000000e+00 1 ## Rasl2-9 4.347557e-04 1.7072993 0.029 0.006 1.000000e+00 1 ## Becn11 4.379610e-04 0.1036891 0.784 0.558 1.000000e+00 1 ## Fam20a1 4.401288e-04 0.3745312 0.233 0.138 1.000000e+00 1 ## Tfpt 4.401761e-04 0.4075580 0.245 0.149 1.000000e+00 1 ## Gdf15 4.456278e-04 0.4578091 0.212 0.122 1.000000e+00 1 ## B3galt6 4.459472e-04 0.4235347 0.196 0.111 1.000000e+00 1 ## Rbm48 4.463300e-04 0.2825093 0.253 0.152 1.000000e+00 1 ## Pctp 4.470999e-04 0.7637296 0.167 0.092 1.000000e+00 1 ## H131 4.505521e-04 0.1113141 0.886 0.672 1.000000e+00 1 ## Tomm61 4.528898e-04 0.1372149 0.918 0.706 1.000000e+00 1 ## 9130401M01Rik 4.542292e-04 0.7552832 0.224 0.137 1.000000e+00 1 ## 4930556M19Rik 4.599193e-04 1.8936239 0.037 0.009 1.000000e+00 1 ## Gm57630 4.599193e-04 1.4857620 0.037 0.009 1.000000e+00 1 ## Klhl15 4.600664e-04 0.2117815 0.204 0.113 1.000000e+00 1 ## St6galnac4 4.654563e-04 0.5640653 0.127 0.062 1.000000e+00 1 ## Gm42012 4.662155e-04 1.8956729 0.037 0.009 1.000000e+00 1 ## Agk 4.697533e-04 0.6384140 0.192 0.113 1.000000e+00 1 ## Pigc 4.718411e-04 0.2844180 0.306 0.196 1.000000e+00 1 ## Mob3b 4.758037e-04 0.2769630 0.367 0.247 1.000000e+00 1 ## Prkaca1 4.769673e-04 0.1155290 0.559 0.373 1.000000e+00 1 ## Gm7457 4.775476e-04 3.1693123 0.020 0.003 1.000000e+00 1 ## Gm4419 4.787614e-04 0.8030380 0.082 0.033 1.000000e+00 1 ## Haus7 4.792727e-04 0.4252057 0.237 0.143 1.000000e+00 1 ## Zfp330 4.820208e-04 0.1364185 0.502 0.331 1.000000e+00 1 ## Pcbp4 4.889758e-04 0.2246348 0.278 0.168 1.000000e+00 1 ## Gm3807 4.895540e-04 0.8860557 0.037 0.009 1.000000e+00 1 ## Zfp119b 4.919946e-04 0.7926705 0.122 0.060 1.000000e+00 1 ## Champ1 4.974197e-04 0.3827586 0.208 0.120 1.000000e+00 1 ## Ldlrap1 4.975996e-04 0.4660560 0.122 0.061 1.000000e+00 1 ## Gm26995 4.981336e-04 2.6641178 0.020 0.003 1.000000e+00 1 ## Zfp9 4.998422e-04 1.9597420 0.041 0.011 1.000000e+00 1 ## LOC102631992 5.002371e-04 1.6683527 0.020 0.003 1.000000e+00 1 ## Gm53773 5.011193e-04 0.6489577 0.086 0.036 1.000000e+00 1 ## Tmem169 5.043480e-04 1.1144693 0.037 0.009 1.000000e+00 1 ## Stub11 5.067402e-04 0.1394677 0.971 0.795 1.000000e+00 1 ## Gm51704 5.083435e-04 2.5888505 0.024 0.004 1.000000e+00 1 ## Gm26844 5.083435e-04 2.2994334 0.024 0.004 1.000000e+00 1 ## Gm52678 5.087342e-04 2.5553738 0.020 0.003 1.000000e+00 1 ## Lrrc73 5.102826e-04 0.8125797 0.098 0.043 1.000000e+00 1 ## Adck2 5.127821e-04 0.3353525 0.257 0.159 1.000000e+00 1 ## Gm46488 5.130329e-04 2.1705960 0.020 0.003 1.000000e+00 1 ## Atp6v0e1 5.185665e-04 0.1310842 0.955 0.761 1.000000e+00 1 ## Snupn 5.218666e-04 0.2848354 0.208 0.122 1.000000e+00 1 ## Ift140 5.258083e-04 0.2609779 0.171 0.096 1.000000e+00 1 ## Prmt5 5.271369e-04 0.2399862 0.314 0.198 1.000000e+00 1 ## Galt 5.291584e-04 0.4279889 0.310 0.205 1.000000e+00 1 ## Gm46195 5.296950e-04 1.3334311 0.057 0.020 1.000000e+00 1 ## Gm39000 5.310570e-04 1.6713339 0.024 0.004 1.000000e+00 1 ## C920021L13Rik 5.343550e-04 0.2392848 0.090 0.038 1.000000e+00 1 ## Bckdha 5.386203e-04 0.2071904 0.400 0.264 1.000000e+00 1 ## Pogz1 5.409042e-04 0.2119971 0.335 0.212 1.000000e+00 1 ## Gm14326 5.431131e-04 0.2766184 0.176 0.099 1.000000e+00 1 ## Nkap 5.466815e-04 0.1351710 0.420 0.279 1.000000e+00 1 ## Memo1 5.481941e-04 0.1979189 0.388 0.256 1.000000e+00 1 ## Fam185a 5.511597e-04 0.6657207 0.163 0.089 1.000000e+00 1 ## Mus81 5.530312e-04 0.9145210 0.122 0.061 1.000000e+00 1 ## Gm46786 5.541023e-04 1.9003158 0.049 0.015 1.000000e+00 1 ## Ube2i1 5.545986e-04 0.1340016 0.910 0.682 1.000000e+00 1 ## Get1 5.546470e-04 1.0709698 0.102 0.047 1.000000e+00 1 ## Tnfrsf14 5.577947e-04 0.1966150 0.273 0.166 1.000000e+00 1 ## Bud23 5.617088e-04 0.1115990 0.449 0.290 1.000000e+00 1 ## Clcn31 5.619382e-04 0.1096738 0.882 0.673 1.000000e+00 1 ## Vps8 5.652240e-04 0.2033786 0.237 0.140 1.000000e+00 1 ## Cep131 5.655608e-04 0.5378376 0.127 0.063 1.000000e+00 1 ## Zfp777 5.659849e-04 0.3405560 0.224 0.133 1.000000e+00 1 ## Gm52861 5.704672e-04 0.5631265 0.082 0.034 1.000000e+00 1 ## Zkscan7 5.729074e-04 0.2656813 0.237 0.141 1.000000e+00 1 ## Dand5 5.740654e-04 0.8266370 0.139 0.072 1.000000e+00 1 ## Cd99 5.750624e-04 1.3012935 0.057 0.020 1.000000e+00 1 ## Stimate 5.768805e-04 0.4177234 0.163 0.089 1.000000e+00 1 ## Trpv6 5.843580e-04 0.5855751 0.045 0.013 1.000000e+00 1 ## AW046200 5.843580e-04 1.7678782 0.045 0.013 1.000000e+00 1 ## Gm43380 5.857526e-04 0.9774292 0.041 0.011 1.000000e+00 1 ## Nudt12 5.858840e-04 0.9919745 0.082 0.034 1.000000e+00 1 ## Zfp41 5.862215e-04 1.0501759 0.069 0.027 1.000000e+00 1 ## Thyn1 5.871566e-04 0.3065173 0.331 0.210 1.000000e+00 1 ## Ngef 5.961727e-04 0.7573596 0.151 0.082 1.000000e+00 1 ## Gm53595 5.963226e-04 0.9744869 0.069 0.027 1.000000e+00 1 ## Rell2 5.983621e-04 1.2453132 0.069 0.027 1.000000e+00 1 ## Gm10614 6.093947e-04 0.8195673 0.049 0.015 1.000000e+00 1 ## Thap1 6.112626e-04 0.3428689 0.212 0.122 1.000000e+00 1 ## Prps1l31 6.157020e-04 0.2625920 0.408 0.272 1.000000e+00 1 ## Ptgds 6.192662e-04 3.3197280 0.012 0.001 1.000000e+00 1 ## 1700026J14Rik 6.192662e-04 3.2525918 0.012 0.001 1.000000e+00 1 ## 4930579G18Rik 6.192662e-04 3.4337434 0.012 0.001 1.000000e+00 1 ## Kcnrg 6.192662e-04 3.3276770 0.012 0.001 1.000000e+00 1 ## Tmem178 6.192662e-04 3.1567128 0.012 0.001 1.000000e+00 1 ## Gm33335 6.192662e-04 3.3800624 0.012 0.001 1.000000e+00 1 ## Akap14 6.192662e-04 3.4852079 0.012 0.001 1.000000e+00 1 ## Alg2 6.208210e-04 0.1130934 0.478 0.323 1.000000e+00 1 ## Gm25663 6.231284e-04 3.3626028 0.012 0.001 1.000000e+00 1 ## Gyg1 6.247206e-04 0.4764711 0.257 0.164 1.000000e+00 1 ## Cyp2d22 6.257148e-04 0.2537095 0.143 0.073 1.000000e+00 1 ## Pcdhga5 6.261887e-04 0.6571769 0.094 0.041 1.000000e+00 1 ## Oaz3 6.270130e-04 3.2894181 0.012 0.001 1.000000e+00 1 ## Cxcl9 6.270130e-04 3.0832779 0.012 0.001 1.000000e+00 1 ## Gm40578 6.270130e-04 3.3665938 0.012 0.001 1.000000e+00 1 ## Gm5215 6.270130e-04 3.2600518 0.012 0.001 1.000000e+00 1 ## Trap1 6.275148e-04 0.2390096 0.384 0.247 1.000000e+00 1 ## Gm34507 6.293429e-04 1.8420435 0.033 0.008 1.000000e+00 1 ## Rbm8a1 6.299725e-04 0.1713089 0.808 0.577 1.000000e+00 1 ## Gm33610 6.309202e-04 2.8717343 0.012 0.001 1.000000e+00 1 ## Gm52486 6.309202e-04 2.8668532 0.012 0.001 1.000000e+00 1 ## Gm53349 6.309202e-04 2.0869534 0.012 0.001 1.000000e+00 1 ## Gm53386 6.309202e-04 2.8403128 0.012 0.001 1.000000e+00 1 ## 4933427E11Rik 6.309202e-04 3.0323804 0.012 0.001 1.000000e+00 1 ## Ltbp3 6.309202e-04 1.9885003 0.012 0.001 1.000000e+00 1 ## Aacs1 6.310917e-04 0.1256894 0.388 0.265 1.000000e+00 1 ## Brf2 6.313850e-04 0.3154972 0.151 0.082 1.000000e+00 1 ## Zfp28 6.317983e-04 0.6139672 0.118 0.057 1.000000e+00 1 ## Fam53a 6.366560e-04 0.2596234 0.388 0.263 1.000000e+00 1 ## D7Ertd128e 6.403830e-04 0.4713963 0.127 0.062 1.000000e+00 1 ## Vps26c1 6.490413e-04 0.1785404 0.453 0.301 1.000000e+00 1 ## Cdin1 6.497574e-04 0.4400674 0.180 0.102 1.000000e+00 1 ## Fbxw81 6.499187e-04 0.3160032 0.359 0.239 1.000000e+00 1 ## Psmd21 6.507221e-04 0.1258205 0.800 0.577 1.000000e+00 1 ## Bcap29 6.528136e-04 0.1832195 0.306 0.196 1.000000e+00 1 ## Ndufv31 6.533258e-04 0.1234593 0.984 0.802 1.000000e+00 1 ## Gaa1 6.563890e-04 0.2575822 0.412 0.284 1.000000e+00 1 ## Qprt 6.566976e-04 1.3887732 0.045 0.013 1.000000e+00 1 ## Map10 6.571102e-04 0.1859907 0.127 0.062 1.000000e+00 1 ## Ipo9 6.579008e-04 0.2023127 0.278 0.173 1.000000e+00 1 ## Gas8 6.589878e-04 0.1748216 0.233 0.139 1.000000e+00 1 ## Pigt1 6.668568e-04 0.1052288 0.702 0.484 1.000000e+00 1 ## Dusp28 6.687514e-04 0.6183454 0.184 0.105 1.000000e+00 1 ## Zfp865 6.703588e-04 0.3605406 0.294 0.187 1.000000e+00 1 ## Zfp658 6.784403e-04 1.2681598 0.049 0.015 1.000000e+00 1 ## Gm6658 6.804543e-04 0.7853536 0.094 0.042 1.000000e+00 1 ## Tbcc1 6.816684e-04 0.3049325 0.506 0.343 1.000000e+00 1 ## Rita1 6.832618e-04 0.5930815 0.135 0.070 1.000000e+00 1 ## Ppox 6.850523e-04 0.3472782 0.237 0.143 1.000000e+00 1 ## Zfp689 6.859405e-04 0.6022328 0.122 0.060 1.000000e+00 1 ## Gm356071 6.872825e-04 0.7891893 0.102 0.047 1.000000e+00 1 ## Fip1l1 6.908507e-04 0.1095991 0.759 0.553 1.000000e+00 1 ## Dnajc9 6.935681e-04 0.2622632 0.298 0.187 1.000000e+00 1 ## Mgat1 6.947439e-04 0.1600591 0.514 0.358 1.000000e+00 1 ## Gmpr2 6.955956e-04 0.2055869 0.261 0.159 1.000000e+00 1 ## Mfhas1 6.991499e-04 0.4151470 0.159 0.087 1.000000e+00 1 ## Gm33409 7.073866e-04 1.7882429 0.033 0.008 1.000000e+00 1 ## Ap4s1 7.149299e-04 0.1950475 0.457 0.310 1.000000e+00 1 ## Tmem218 7.168805e-04 0.7262054 0.147 0.080 1.000000e+00 1 ## Gm14051 7.174964e-04 1.5232739 0.033 0.008 1.000000e+00 1 ## Rnaseh2a 7.201421e-04 0.1401434 0.290 0.177 1.000000e+00 1 ## Hoxb2 7.202432e-04 0.5420782 0.094 0.042 1.000000e+00 1 ## Cdc42ep41 7.236626e-04 0.1367250 0.502 0.340 1.000000e+00 1 ## Prkd2 7.266962e-04 0.2522806 0.216 0.129 1.000000e+00 1 ## Pex26 7.294584e-04 0.2391152 0.314 0.204 1.000000e+00 1 ## Ttll1 7.299777e-04 0.3699807 0.159 0.085 1.000000e+00 1 ## Hdac21 7.347672e-04 0.1492733 0.616 0.446 1.000000e+00 1 ## Coa4 7.393319e-04 0.2272831 0.380 0.253 1.000000e+00 1 ## Dmap11 7.476902e-04 0.1788960 0.359 0.236 1.000000e+00 1 ## Tpgs2 7.508671e-04 0.6839352 0.135 0.071 1.000000e+00 1 ## Gmppa 7.563380e-04 0.1208683 0.580 0.412 1.000000e+00 1 ## Phykpl 7.647256e-04 0.2274851 0.294 0.189 1.000000e+00 1 ## Kpna61 7.648262e-04 0.1157584 0.424 0.280 1.000000e+00 1 ## Selenbp1 7.774572e-04 0.1218747 0.204 0.122 1.000000e+00 1 ## Thumpd3 7.782217e-04 0.2675925 0.322 0.210 1.000000e+00 1 ## Ldhd 7.855840e-04 1.1392075 0.078 0.033 1.000000e+00 1 ## Nfatc2 7.952525e-04 0.5371105 0.114 0.057 1.000000e+00 1 ## Mir703 7.955317e-04 0.2686254 0.331 0.211 1.000000e+00 1 ## Gtpbp8 7.990790e-04 0.2631650 0.257 0.161 1.000000e+00 1 ## 2210417A02Rik1 8.063847e-04 0.6143365 0.094 0.042 1.000000e+00 1 ## Mnd1 8.087887e-04 1.4490156 0.078 0.033 1.000000e+00 1 ## Slc19a2 8.104341e-04 0.5579092 0.192 0.110 1.000000e+00 1 ## Rela 8.117611e-04 0.3922730 0.314 0.208 1.000000e+00 1 ## Pemt 8.134844e-04 1.0075426 0.127 0.066 1.000000e+00 1 ## Pcsk2 8.173478e-04 0.6573726 0.216 0.133 1.000000e+00 1 ## Pum3 8.201863e-04 0.1775820 0.416 0.277 1.000000e+00 1 ## Acp11 8.249111e-04 0.1120078 0.800 0.585 1.000000e+00 1 ## Gm40448 8.284926e-04 1.7556697 0.041 0.012 1.000000e+00 1 ## Gm46941 8.295673e-04 1.8492824 0.037 0.010 1.000000e+00 1 ## Zkscan17 8.301439e-04 0.6208695 0.167 0.094 1.000000e+00 1 ## 9330020H09Rik 8.364213e-04 0.2591205 0.220 0.129 1.000000e+00 1 ## 2010001A14Rik 8.490479e-04 0.2934294 0.261 0.165 1.000000e+00 1 ## Gm41408 8.499908e-04 0.8376089 0.078 0.032 1.000000e+00 1 ## Gosr1 8.585447e-04 0.1658719 0.371 0.247 1.000000e+00 1 ## Zfp1101 8.632889e-04 0.1759728 0.437 0.304 1.000000e+00 1 ## Nsdhl 8.635267e-04 0.1860745 0.261 0.161 1.000000e+00 1 ## Zfp39 8.682925e-04 0.8345123 0.057 0.020 1.000000e+00 1 ## Reep2 8.751959e-04 1.4392368 0.037 0.010 1.000000e+00 1 ## Kdm2b 8.825711e-04 0.4016942 0.298 0.194 1.000000e+00 1 ## Ribc1 8.830521e-04 0.9924052 0.065 0.025 1.000000e+00 1 ## Parp3 8.878515e-04 0.9176407 0.094 0.043 1.000000e+00 1 ## Gas2l3 8.993491e-04 0.2637664 0.110 0.054 1.000000e+00 1 ## Gm46775 8.997591e-04 0.9346589 0.053 0.018 1.000000e+00 1 ## Ak6 9.005594e-04 0.1799336 0.424 0.286 1.000000e+00 1 ## 6430503K07Rik 9.015350e-04 0.5268377 0.110 0.052 1.000000e+00 1 ## Purg 9.084813e-04 0.4820494 0.122 0.061 1.000000e+00 1 ## Bloc1s3 9.112950e-04 0.4258516 0.155 0.083 1.000000e+00 1 ## Gm10087 9.123880e-04 0.5906845 0.041 0.012 1.000000e+00 1 ## Zbtb34 9.135608e-04 0.3141340 0.127 0.064 1.000000e+00 1 ## Lrfn1 9.140313e-04 2.2742162 0.029 0.006 1.000000e+00 1 ## Osbpl8 9.152561e-04 0.4143826 0.208 0.123 1.000000e+00 1 ## Pdp2 9.186415e-04 0.1248590 0.327 0.212 1.000000e+00 1 ## Dnajc16 9.187948e-04 0.3593050 0.176 0.099 1.000000e+00 1 ## Rbm31 9.206331e-04 0.1290288 0.559 0.375 1.000000e+00 1 ## Ogdhl 9.214209e-04 0.4743972 0.151 0.081 1.000000e+00 1 ## Tmem184c 9.233591e-04 0.2142583 0.322 0.208 1.000000e+00 1 ## Pcnx4 9.235427e-04 0.1376998 0.261 0.159 1.000000e+00 1 ## Fnbp1 9.239686e-04 0.1377160 0.265 0.161 1.000000e+00 1 ## Tmem91 9.364753e-04 2.0484412 0.029 0.006 1.000000e+00 1 ## Katnb1 9.379981e-04 0.6822345 0.155 0.087 1.000000e+00 1 ## Map6 9.394088e-04 0.3140454 0.220 0.131 1.000000e+00 1 ## Gm8290 9.401097e-04 1.4301294 0.049 0.016 1.000000e+00 1 ## Carmil2 9.452587e-04 1.0564185 0.065 0.025 1.000000e+00 1 ## Epb42 9.533138e-04 1.2555162 0.065 0.025 1.000000e+00 1 ## Gm41845 9.536422e-04 1.9388062 0.029 0.006 1.000000e+00 1 ## Tctn1 9.559001e-04 0.7090277 0.090 0.041 1.000000e+00 1 ## Gm4532 9.567216e-04 0.2250805 0.106 0.050 1.000000e+00 1 ## Aars2 9.578197e-04 0.6310272 0.184 0.107 1.000000e+00 1 ## Mast1 9.583452e-04 1.3649119 0.041 0.012 1.000000e+00 1 ## Prmt7 9.584659e-04 0.3973902 0.237 0.145 1.000000e+00 1 ## Clybl 9.586525e-04 0.4385562 0.204 0.124 1.000000e+00 1 ## Cd247 9.628321e-04 1.3628424 0.041 0.012 1.000000e+00 1 ## Zfp647 9.695990e-04 1.3588178 0.041 0.012 1.000000e+00 1 ## Rsph4a 9.740385e-04 1.5656315 0.029 0.006 1.000000e+00 1 ## Ppp2r5b1 9.773179e-04 0.1727108 0.522 0.379 1.000000e+00 1 ## 5730409E04Rik 9.829028e-04 1.9845273 0.029 0.006 1.000000e+00 1 ## Eipr1 9.839810e-04 0.1947922 0.318 0.205 1.000000e+00 1 ## Cpsf3 9.914775e-04 0.1440513 0.404 0.268 1.000000e+00 1 ## Fgd1 9.948746e-04 0.8814919 0.098 0.046 1.000000e+00 1 ## Fam199x1 1.001545e-03 0.2019851 0.392 0.269 1.000000e+00 1 ## Svop 1.004691e-03 0.9093771 0.065 0.025 1.000000e+00 1 ## Pcdhga6 1.011504e-03 0.5504585 0.065 0.025 1.000000e+00 1 ## Pbx2 1.015456e-03 0.1457345 0.441 0.304 1.000000e+00 1 ## Gm5296 1.016644e-03 0.6029111 0.171 0.099 1.000000e+00 1 ## Prcp 1.020114e-03 0.7248919 0.163 0.092 1.000000e+00 1 ## AB010352 1.036567e-03 0.2002171 0.204 0.119 1.000000e+00 1 ## Septin9 1.039809e-03 0.3710989 0.302 0.200 1.000000e+00 1 ## H2ap 1.041038e-03 1.3571345 0.045 0.014 1.000000e+00 1 ## Tmem68 1.041691e-03 0.2316061 0.286 0.181 1.000000e+00 1 ## Ubfd1 1.055056e-03 0.1349445 0.535 0.370 1.000000e+00 1 ## Ppm1g1 1.059258e-03 0.1666683 0.584 0.411 1.000000e+00 1 ## R3hcc1 1.062953e-03 0.3625290 0.163 0.093 1.000000e+00 1 ## 2010315B03Rik 1.064984e-03 0.2285771 0.335 0.225 1.000000e+00 1 ## Gm16845 1.075716e-03 0.9318498 0.090 0.041 1.000000e+00 1 ## Dcaf12l1 1.077405e-03 0.6742442 0.073 0.030 1.000000e+00 1 ## Kif3c 1.077498e-03 0.2553531 0.180 0.100 1.000000e+00 1 ## Fundc1 1.077707e-03 0.4196009 0.273 0.176 1.000000e+00 1 ## Tmem185b1 1.078734e-03 0.3273592 0.233 0.143 1.000000e+00 1 ## Megf8 1.081759e-03 0.1555285 0.237 0.142 1.000000e+00 1 ## Snhg91 1.091134e-03 0.1153655 0.355 0.234 1.000000e+00 1 ## Gm5914 1.095713e-03 0.4941191 0.237 0.147 1.000000e+00 1 ## Klhl5 1.101591e-03 1.3087996 0.082 0.036 1.000000e+00 1 ## Brsk1 1.108328e-03 0.3385463 0.143 0.075 1.000000e+00 1 ## Zfp582 1.116328e-03 0.9796335 0.061 0.023 1.000000e+00 1 ## Bbs7 1.120714e-03 0.7639307 0.098 0.046 1.000000e+00 1 ## Catsper2 1.123176e-03 0.7942340 0.102 0.049 1.000000e+00 1 ## Invs 1.136447e-03 0.6653481 0.147 0.081 1.000000e+00 1 ## Parp11 1.136687e-03 0.3804915 0.176 0.101 1.000000e+00 1 ## Crlf3 1.144436e-03 0.3240002 0.208 0.128 1.000000e+00 1 ## Meaf61 1.148364e-03 0.2201822 0.522 0.368 1.000000e+00 1 ## 1600012H06Rik 1.154552e-03 0.1454614 0.367 0.236 1.000000e+00 1 ## Rnaseh1 1.182503e-03 0.4036904 0.220 0.136 1.000000e+00 1 ## Isy1 1.186613e-03 0.1785028 0.249 0.154 1.000000e+00 1 ## Gm40190 1.192660e-03 1.2210358 0.057 0.021 1.000000e+00 1 ## Bicdl2 1.193960e-03 0.4038712 0.224 0.138 1.000000e+00 1 ## Cdyl 1.195862e-03 0.2816961 0.241 0.150 1.000000e+00 1 ## Gm30706 1.203494e-03 0.9892463 0.057 0.021 1.000000e+00 1 ## Mex3c 1.206666e-03 0.1155362 0.514 0.356 1.000000e+00 1 ## Mrps111 1.209715e-03 0.1182675 0.506 0.351 1.000000e+00 1 ## Atp5f1e1 1.214765e-03 0.1100081 1.000 0.887 1.000000e+00 1 ## Zfp583 1.216611e-03 0.5278883 0.106 0.051 1.000000e+00 1 ## Tlnrd11 1.219253e-03 0.1307597 0.669 0.487 1.000000e+00 1 ## Zscan25 1.222177e-03 0.1461497 0.171 0.096 1.000000e+00 1 ## Dipk1b 1.227807e-03 1.5804083 0.053 0.019 1.000000e+00 1 ## Mfsd14b1 1.228724e-03 0.1141956 0.502 0.338 1.000000e+00 1 ## Gm52692 1.260486e-03 0.9833271 0.024 0.005 1.000000e+00 1 ## Il17ra 1.262145e-03 0.1552728 0.257 0.160 1.000000e+00 1 ## Hps6 1.278803e-03 0.5540998 0.122 0.063 1.000000e+00 1 ## Gm22063 1.287459e-03 1.3983377 0.033 0.008 1.000000e+00 1 ## Chn11 1.308958e-03 1.0076072 0.065 0.026 1.000000e+00 1 ## Plp1 1.309912e-03 0.1782159 0.102 0.048 1.000000e+00 1 ## Zfp105 1.326489e-03 0.9087438 0.053 0.019 1.000000e+00 1 ## Or5m3b 1.328985e-03 1.2858798 0.053 0.019 1.000000e+00 1 ## Gm32449 1.343102e-03 1.4836498 0.033 0.008 1.000000e+00 1 ## Dmkn 1.344052e-03 0.5525115 0.053 0.019 1.000000e+00 1 ## Ppih 1.346400e-03 0.1877328 0.290 0.182 1.000000e+00 1 ## Casp9 1.349624e-03 0.3706267 0.220 0.134 1.000000e+00 1 ## Slc2a8 1.349643e-03 0.5580823 0.159 0.091 1.000000e+00 1 ## Pstk 1.356118e-03 0.4986499 0.184 0.113 1.000000e+00 1 ## Gm22299 1.357356e-03 1.5803309 0.033 0.008 1.000000e+00 1 ## Tjap1 1.367295e-03 0.2636697 0.278 0.177 1.000000e+00 1 ## Bloc1s5 1.369898e-03 0.5570491 0.216 0.135 1.000000e+00 1 ## Gm15927 1.383153e-03 0.7692770 0.078 0.034 1.000000e+00 1 ## Gm33315 1.390364e-03 1.9296866 0.041 0.013 1.000000e+00 1 ## Herc6 1.398339e-03 0.1802403 0.151 0.082 1.000000e+00 1 ## Mrpl39 1.399007e-03 0.1881912 0.322 0.209 1.000000e+00 1 ## Kcnq4 1.399104e-03 3.0625887 0.016 0.002 1.000000e+00 1 ## Zfp606 1.401866e-03 0.1084288 0.322 0.211 1.000000e+00 1 ## Eif2ak4 1.409739e-03 0.6217144 0.122 0.064 1.000000e+00 1 ## Avp 1.411554e-03 2.9339168 0.016 0.002 1.000000e+00 1 ## Gm38529 1.411687e-03 0.1942792 0.078 0.034 1.000000e+00 1 ## Adamtsl4 1.417818e-03 2.4978876 0.016 0.002 1.000000e+00 1 ## Lrrc8c 1.422594e-03 1.3860893 0.037 0.010 1.000000e+00 1 ## Kmo 1.424107e-03 2.6596609 0.016 0.002 1.000000e+00 1 ## Gm33782 1.424107e-03 2.7878699 0.016 0.002 1.000000e+00 1 ## Eno1b 1.425952e-03 1.5990051 0.045 0.015 1.000000e+00 1 ## Gm13032 1.426015e-03 1.7975982 0.037 0.010 1.000000e+00 1 ## Cd72 1.430422e-03 2.7794229 0.016 0.002 1.000000e+00 1 ## Prkab1 1.432840e-03 0.2487432 0.294 0.196 1.000000e+00 1 ## Acbd7 1.436762e-03 1.7808677 0.016 0.002 1.000000e+00 1 ## E130112N10Rik 1.436762e-03 1.9785170 0.016 0.002 1.000000e+00 1 ## Gm53491 1.449521e-03 2.1559184 0.016 0.002 1.000000e+00 1 ## Zgpat 1.455376e-03 0.1358887 0.237 0.147 1.000000e+00 1 ## 4930448E22Rik 1.455940e-03 1.8860804 0.016 0.002 1.000000e+00 1 ## Pheta2 1.455940e-03 1.5715490 0.016 0.002 1.000000e+00 1 ## Gm29863 1.455940e-03 2.5817638 0.016 0.002 1.000000e+00 1 ## Fam89b1 1.458066e-03 0.1741850 0.408 0.277 1.000000e+00 1 ## Spsb21 1.460297e-03 0.1426222 0.367 0.244 1.000000e+00 1 ## Adnp 1.466842e-03 0.1920813 0.224 0.138 1.000000e+00 1 ## Gm54256 1.476469e-03 1.1070924 0.041 0.013 1.000000e+00 1 ## Gm15478 1.478956e-03 0.7318109 0.078 0.034 1.000000e+00 1 ## Tmeff1 1.483569e-03 0.1277468 0.314 0.203 1.000000e+00 1 ## Prcc 1.483814e-03 0.1460881 0.298 0.198 1.000000e+00 1 ## 9030612E09Rik 1.485366e-03 1.0941343 0.049 0.017 1.000000e+00 1 ## Zfp783 1.486202e-03 1.3331356 0.065 0.027 1.000000e+00 1 ## Ggnbp1 1.496589e-03 0.7800094 0.069 0.029 1.000000e+00 1 ## Snhg14 1.498222e-03 1.5184184 0.057 0.022 1.000000e+00 1 ## Gm35374 1.501049e-03 2.2693965 0.020 0.003 1.000000e+00 1 ## Mtnr1a 1.506598e-03 2.4368726 0.020 0.003 1.000000e+00 1 ## Zfp1006 1.510257e-03 0.4328708 0.143 0.078 1.000000e+00 1 ## Wdr31 1.515447e-03 0.7084173 0.082 0.036 1.000000e+00 1 ## Tub 1.517741e-03 0.9066097 0.049 0.017 1.000000e+00 1 ## Agbl5 1.527816e-03 0.1063880 0.167 0.094 1.000000e+00 1 ## Iqch 1.534627e-03 2.2776212 0.020 0.003 1.000000e+00 1 ## Gm52500 1.540291e-03 2.1509502 0.020 0.003 1.000000e+00 1 ## Klhl25 1.540464e-03 0.9489467 0.102 0.050 1.000000e+00 1 ## Stambpl1 1.558418e-03 0.2660174 0.208 0.129 1.000000e+00 1 ## Gm19744 1.563136e-03 1.3225660 0.020 0.003 1.000000e+00 1 ## Gm31434 1.564110e-03 0.2548168 0.102 0.049 1.000000e+00 1 ## Vrk2 1.572258e-03 1.0481125 0.094 0.045 1.000000e+00 1 ## Dcun1d3 1.589183e-03 0.4184741 0.180 0.105 1.000000e+00 1 ## Metap21 1.626386e-03 0.1415635 0.984 0.784 1.000000e+00 1 ## Ncoa5 1.628236e-03 0.2624494 0.384 0.268 1.000000e+00 1 ## Acyp2 1.659975e-03 0.2626481 0.114 0.058 1.000000e+00 1 ## Nr1h3 1.678736e-03 0.2272716 0.273 0.180 1.000000e+00 1 ## Pet117 1.688192e-03 0.2519266 0.180 0.106 1.000000e+00 1 ## Ddx47 1.689611e-03 0.1009140 0.335 0.220 1.000000e+00 1 ## 2700049A03Rik 1.690313e-03 0.4847625 0.180 0.107 1.000000e+00 1 ## Gm47340 1.700536e-03 0.9384313 0.094 0.045 1.000000e+00 1 ## Srf1 1.701037e-03 0.1203381 0.371 0.242 1.000000e+00 1 ## Trpt1 1.708151e-03 0.9869429 0.086 0.040 1.000000e+00 1 ## Eme2 1.717209e-03 1.0865602 0.106 0.054 1.000000e+00 1 ## Fitm2 1.733064e-03 0.9255676 0.065 0.027 1.000000e+00 1 ## Cluap1 1.733336e-03 0.2321530 0.294 0.192 1.000000e+00 1 ## Urm1 1.737582e-03 0.1656249 0.424 0.290 1.000000e+00 1 ## Mib2 1.746829e-03 0.1163894 0.310 0.203 1.000000e+00 1 ## Rab40b 1.760866e-03 0.8198153 0.078 0.034 1.000000e+00 1 ## Mterf1b 1.766237e-03 0.2698883 0.176 0.101 1.000000e+00 1 ## Arf2 1.771010e-03 0.1825867 0.212 0.131 1.000000e+00 1 ## Pinlyp 1.784971e-03 2.5294242 0.029 0.007 1.000000e+00 1 ## Nceh1 1.793828e-03 0.3449114 0.180 0.106 1.000000e+00 1 ## Lrrc49 1.794813e-03 0.3001428 0.171 0.100 1.000000e+00 1 ## Gm3363 1.794983e-03 2.4113380 0.029 0.007 1.000000e+00 1 ## Ndufaf1 1.795736e-03 0.4467941 0.192 0.118 1.000000e+00 1 ## Emc11 1.812691e-03 0.1210416 0.449 0.311 1.000000e+00 1 ## Sav1 1.832338e-03 0.1167682 0.400 0.284 1.000000e+00 1 ## Cbr3 1.840681e-03 0.3206497 0.029 0.007 1.000000e+00 1 ## Vps51 1.854831e-03 0.1243290 0.335 0.223 1.000000e+00 1 ## Wdr93 1.861332e-03 1.7805191 0.029 0.007 1.000000e+00 1 ## Tekt41 1.871794e-03 0.4317889 0.151 0.082 1.000000e+00 1 ## Zdhhc6 1.874897e-03 0.1650989 0.347 0.234 1.000000e+00 1 ## F830010H11Rik 1.876960e-03 1.5643819 0.029 0.007 1.000000e+00 1 ## Zfp790 1.880824e-03 0.2724239 0.265 0.175 1.000000e+00 1 ## Fdxr 1.888128e-03 0.3838906 0.196 0.120 1.000000e+00 1 ## Pcdhgb2 1.919225e-03 1.2797358 0.029 0.007 1.000000e+00 1 ## Fam151b 1.936478e-03 0.7672390 0.098 0.048 1.000000e+00 1 ## Rgs6 1.938366e-03 0.1158632 0.057 0.022 1.000000e+00 1 ## Gm11723 1.940684e-03 1.1449630 0.029 0.007 1.000000e+00 1 ## Guca1a 1.941949e-03 0.9646523 0.098 0.049 1.000000e+00 1 ## Mccc1 1.943922e-03 0.1282445 0.306 0.203 1.000000e+00 1 ## Zfp346 1.962659e-03 0.3013389 0.143 0.078 1.000000e+00 1 ## Gm2885 1.967818e-03 1.5267122 0.029 0.007 1.000000e+00 1 ## Usb1 1.971892e-03 0.5329399 0.176 0.104 1.000000e+00 1 ## Inpp5b 1.976007e-03 0.1177124 0.261 0.171 1.000000e+00 1 ## Dnttip21 1.981905e-03 0.1189320 0.641 0.461 1.000000e+00 1 ## Snx16 1.984710e-03 0.1619208 0.327 0.219 1.000000e+00 1 ## Pofut1 1.998061e-03 0.3229904 0.224 0.141 1.000000e+00 1 ## 1110002L01Rik 2.013608e-03 0.1181171 0.233 0.141 1.000000e+00 1 ## Gm41774 2.024040e-03 1.5935023 0.049 0.017 1.000000e+00 1 ## Ssh1 2.024640e-03 0.1628737 0.351 0.236 1.000000e+00 1 ## Zdhhc4 2.037334e-03 0.2303221 0.265 0.171 1.000000e+00 1 ## Nol11 2.052375e-03 0.2683028 0.347 0.230 1.000000e+00 1 ## Gatb 2.063726e-03 0.6942661 0.151 0.088 1.000000e+00 1 ## Wdtc11 2.072701e-03 0.1429034 0.441 0.300 1.000000e+00 1 ## Gm53858 2.078842e-03 2.2915077 0.033 0.009 1.000000e+00 1 ## Ogfod1 2.101552e-03 0.1155602 0.261 0.163 1.000000e+00 1 ## Phldb3 2.105609e-03 0.3512420 0.122 0.065 1.000000e+00 1 ## Gpsm1 2.109136e-03 0.1400068 0.163 0.092 1.000000e+00 1 ## Erich5 2.147108e-03 1.9746936 0.033 0.009 1.000000e+00 1 ## Wdr44 2.151518e-03 0.1309609 0.343 0.234 1.000000e+00 1 ## Fmnl1 2.154101e-03 0.8228998 0.073 0.032 1.000000e+00 1 ## Mrtfa 2.157842e-03 0.1222434 0.302 0.195 1.000000e+00 1 ## Lrrc51 2.158574e-03 0.5108515 0.114 0.059 1.000000e+00 1 ## Mbip 2.181338e-03 0.3115998 0.208 0.129 1.000000e+00 1 ## Twf21 2.183742e-03 0.2044476 0.339 0.227 1.000000e+00 1 ## AW554918 2.193522e-03 0.3252338 0.151 0.087 1.000000e+00 1 ## Fbh11 2.205864e-03 0.1257428 0.482 0.343 1.000000e+00 1 ## Gm35315 2.217423e-03 1.7206847 0.033 0.009 1.000000e+00 1 ## Gm57533 2.222919e-03 1.1283372 0.033 0.009 1.000000e+00 1 ## Gm5785 2.243255e-03 0.1367786 0.171 0.101 1.000000e+00 1 ## C330016L05Rik 2.261737e-03 1.7329855 0.033 0.009 1.000000e+00 1 ## Gm16740 2.262740e-03 0.9762505 0.082 0.038 1.000000e+00 1 ## C030006K11Rik 2.270296e-03 0.5373643 0.151 0.087 1.000000e+00 1 ## Mtmr12 2.278955e-03 0.3873803 0.216 0.138 1.000000e+00 1 ## 4930458D05Rik 2.283915e-03 1.3179954 0.041 0.013 1.000000e+00 1 ## Nckap5l 2.312559e-03 1.5910737 0.033 0.009 1.000000e+00 1 ## 1700088E04Rik 2.312961e-03 0.4710471 0.122 0.065 1.000000e+00 1 ## Brat1 2.316438e-03 0.6304083 0.139 0.078 1.000000e+00 1 ## Abraxas2 2.322061e-03 0.1150588 0.384 0.256 1.000000e+00 1 ## Zfp629 2.324249e-03 0.1253054 0.245 0.157 1.000000e+00 1 ## Lrig2 2.333545e-03 0.2718571 0.233 0.146 1.000000e+00 1 ## Tysnd1 2.342370e-03 0.3772438 0.261 0.175 1.000000e+00 1 ## Vav21 2.364629e-03 0.1739065 0.261 0.169 1.000000e+00 1 ## Scly 2.365471e-03 0.1490117 0.404 0.279 1.000000e+00 1 ## Sh2b3 2.371738e-03 0.2460166 0.269 0.175 1.000000e+00 1 ## Polr1e 2.382009e-03 0.7708122 0.131 0.073 1.000000e+00 1 ## Nme7 2.399305e-03 0.4173419 0.180 0.109 1.000000e+00 1 ## Abca5 2.406214e-03 0.2064635 0.208 0.129 1.000000e+00 1 ## Gcfc2 2.406367e-03 0.6239380 0.131 0.072 1.000000e+00 1 ## Pax41 2.413548e-03 0.2762769 0.171 0.101 1.000000e+00 1 ## Znhit6 2.419466e-03 0.2924823 0.282 0.182 1.000000e+00 1 ## Syngr1 2.465414e-03 0.6060016 0.110 0.057 1.000000e+00 1 ## Tmem198b 2.496319e-03 1.0837551 0.086 0.041 1.000000e+00 1 ## Gm14023 2.504092e-03 0.9590093 0.053 0.020 1.000000e+00 1 ## Zfp229 2.513832e-03 0.5534101 0.127 0.068 1.000000e+00 1 ## Slc6a4 2.538386e-03 0.1894468 0.204 0.129 1.000000e+00 1 ## Uxt1 2.572153e-03 0.1982258 0.420 0.294 1.000000e+00 1 ## Gm20346 2.580557e-03 0.4298400 0.241 0.157 1.000000e+00 1 ## Rhno1 2.581127e-03 0.1468411 0.322 0.212 1.000000e+00 1 ## Zfp768 2.585429e-03 0.3433910 0.233 0.147 1.000000e+00 1 ## Crppa 2.593358e-03 0.6044209 0.102 0.052 1.000000e+00 1 ## Riok2 2.601610e-03 0.1211350 0.261 0.170 1.000000e+00 1 ## Timp2 2.609619e-03 0.5905758 0.127 0.069 1.000000e+00 1 ## Otub2 2.611088e-03 1.3952791 0.037 0.011 1.000000e+00 1 ## Agl 2.648725e-03 0.2462310 0.212 0.131 1.000000e+00 1 ## Rap1b1 2.648781e-03 0.1088982 0.967 0.808 1.000000e+00 1 ## Sema4c 2.665337e-03 0.5895120 0.098 0.048 1.000000e+00 1 ## Zfp526 2.667112e-03 0.3425842 0.094 0.046 1.000000e+00 1 ## Gm154171 2.673941e-03 0.1932188 0.376 0.256 1.000000e+00 1 ## Kbtbd13 2.682417e-03 2.2405762 0.024 0.006 1.000000e+00 1 ## Klhl8 2.685139e-03 0.7265114 0.106 0.055 1.000000e+00 1 ## Homez 2.689647e-03 0.2081458 0.163 0.095 1.000000e+00 1 ## Rbm3os 2.730723e-03 2.5607807 0.024 0.006 1.000000e+00 1 ## Mtr 2.741269e-03 2.1980809 0.045 0.016 1.000000e+00 1 ## Sephs1 2.759528e-03 0.2206018 0.327 0.220 1.000000e+00 1 ## Trib3 2.771601e-03 0.4823508 0.127 0.068 1.000000e+00 1 ## Atg71 2.790174e-03 0.1988520 0.229 0.145 1.000000e+00 1 ## Dhrs13os 2.796368e-03 1.7967817 0.024 0.006 1.000000e+00 1 ## Zfp599 2.798402e-03 1.0150604 0.094 0.048 1.000000e+00 1 ## Phc21 2.802291e-03 0.1057177 0.620 0.454 1.000000e+00 1 ## Bcl2l13 2.822310e-03 0.1529823 0.318 0.216 1.000000e+00 1 ## Nudt8 2.835226e-03 0.3092704 0.286 0.192 1.000000e+00 1 ## Tmem199 2.842464e-03 0.1292780 0.408 0.287 1.000000e+00 1 ## Mycl 2.883415e-03 0.3443752 0.167 0.101 1.000000e+00 1 ## Osbpl7 2.884966e-03 0.2429956 0.220 0.140 1.000000e+00 1 ## Fcf11 2.885953e-03 0.1248148 0.400 0.277 1.000000e+00 1 ## Alkbh3 2.902732e-03 0.3465737 0.286 0.193 1.000000e+00 1 ## Zfp46 2.927516e-03 0.2026171 0.167 0.098 1.000000e+00 1 ## Paxip11 2.949177e-03 0.1712211 0.335 0.226 1.000000e+00 1 ## Rpp30 2.950668e-03 0.5764110 0.200 0.129 1.000000e+00 1 ## Hcn2 2.994592e-03 0.1667530 0.147 0.083 1.000000e+00 1 ## Msl3l21 3.005709e-03 0.2298349 0.090 0.043 1.000000e+00 1 ## Pdp1 3.039196e-03 1.0354004 0.057 0.023 1.000000e+00 1 ## Ripk2 3.041560e-03 0.6410065 0.090 0.044 1.000000e+00 1 ## Ppp1r3e 3.053524e-03 0.6835039 0.078 0.036 1.000000e+00 1 ## Ghdc 3.064987e-03 0.4116045 0.176 0.107 1.000000e+00 1 ## Arl16 3.086909e-03 0.2111218 0.269 0.180 1.000000e+00 1 ## Zfp940 3.087435e-03 0.9031511 0.049 0.018 1.000000e+00 1 ## Gm31834 3.087435e-03 1.2429276 0.049 0.018 1.000000e+00 1 ## Phtf1 3.097620e-03 0.2072309 0.265 0.173 1.000000e+00 1 ## Gm20939 3.099862e-03 0.5148414 0.086 0.041 1.000000e+00 1 ## Slc26a10 3.149046e-03 0.8261864 0.049 0.018 1.000000e+00 1 ## 2510003B16Rik 3.161626e-03 1.0979825 0.069 0.031 1.000000e+00 1 ## Gm41776 3.174197e-03 0.7959664 0.078 0.036 1.000000e+00 1 ## Padi1 3.193729e-03 0.5647525 0.110 0.058 1.000000e+00 1 ## 5330438D12Rik 3.215914e-03 0.2887204 0.122 0.065 1.000000e+00 1 ## Ndufaf5 3.228402e-03 0.4543156 0.196 0.124 1.000000e+00 1 ## Gm29938 3.236101e-03 0.3888821 0.135 0.074 1.000000e+00 1 ## Rab28 3.244042e-03 0.2543411 0.424 0.303 1.000000e+00 1 ## Zfp524 3.254459e-03 0.2312726 0.331 0.227 1.000000e+00 1 ## St3gal1 3.270808e-03 0.4770791 0.094 0.047 1.000000e+00 1 ## Itfg21 3.278336e-03 0.1827020 0.306 0.205 1.000000e+00 1 ## Rbbp9 3.312576e-03 0.4648871 0.171 0.104 1.000000e+00 1 ## Gm461181 3.328756e-03 0.3879986 0.049 0.018 1.000000e+00 1 ## Dpysl5 3.345498e-03 0.8741271 0.073 0.034 1.000000e+00 1 ## Zfp1007 3.360338e-03 0.3861383 0.139 0.078 1.000000e+00 1 ## Exoc3l 3.366021e-03 1.3101952 0.045 0.016 1.000000e+00 1 ## Gm40218 3.380647e-03 0.9104334 0.061 0.026 1.000000e+00 1 ## Gm54308 3.386065e-03 1.9077086 0.029 0.008 1.000000e+00 1 ## Rilpl1 3.393847e-03 0.8935416 0.041 0.014 1.000000e+00 1 ## 1700086P04Rik 3.410676e-03 1.0064043 0.045 0.016 1.000000e+00 1 ## Gon7 3.415005e-03 0.2307116 0.294 0.200 1.000000e+00 1 ## Zfp532 3.442275e-03 0.6103469 0.082 0.039 1.000000e+00 1 ## Lyrm7 3.493388e-03 0.6603246 0.135 0.077 1.000000e+00 1 ## Wnt2b 3.519195e-03 1.1976916 0.029 0.008 1.000000e+00 1 ## Gm41638 3.573413e-03 1.8167684 0.033 0.010 1.000000e+00 1 ## Ubxn81 3.597327e-03 0.1030451 0.327 0.219 1.000000e+00 1 ## AI429214 3.624888e-03 1.1559720 0.041 0.014 1.000000e+00 1 ## Abcb11 3.629109e-03 0.4564637 0.029 0.008 1.000000e+00 1 ## Armcx6 3.632107e-03 1.2470595 0.041 0.014 1.000000e+00 1 ## Shisa51 3.640609e-03 0.1257281 0.763 0.597 1.000000e+00 1 ## Gm53993 3.659669e-03 2.5415963 0.020 0.004 1.000000e+00 1 ## Isca11 3.670875e-03 0.1147268 0.673 0.497 1.000000e+00 1 ## Gm16023 3.683140e-03 0.5708731 0.086 0.042 1.000000e+00 1 ## Sass6 3.683546e-03 0.2817844 0.180 0.109 1.000000e+00 1 ## Gm54192 3.695671e-03 2.4101493 0.020 0.004 1.000000e+00 1 ## Ank1 3.707744e-03 1.8162189 0.020 0.004 1.000000e+00 1 ## Gm53893 3.716969e-03 1.2721908 0.033 0.010 1.000000e+00 1 ## Ap1s2 3.724358e-03 0.3383815 0.143 0.082 1.000000e+00 1 ## Gm53774 3.744175e-03 1.7483785 0.020 0.004 1.000000e+00 1 ## Gm35946 3.768643e-03 2.2348965 0.020 0.004 1.000000e+00 1 ## Klhl21 3.789948e-03 0.1208505 0.473 0.331 1.000000e+00 1 ## Tbx3os11 3.812924e-03 0.2056104 0.171 0.100 1.000000e+00 1 ## Sirt4 3.825468e-03 0.3973530 0.200 0.125 1.000000e+00 1 ## Uba6 3.828293e-03 0.1597088 0.278 0.180 1.000000e+00 1 ## Zfp169 3.831331e-03 0.2019298 0.184 0.111 1.000000e+00 1 ## Ift56 3.845656e-03 0.2651893 0.102 0.052 1.000000e+00 1 ## G630018N14Rik 3.847968e-03 1.1978221 0.033 0.010 1.000000e+00 1 ## Fto 3.864493e-03 0.1372530 0.269 0.180 1.000000e+00 1 ## Mogat1 3.867976e-03 1.6756012 0.020 0.004 1.000000e+00 1 ## Smg8 3.870027e-03 0.3382987 0.159 0.094 1.000000e+00 1 ## Vps45 3.870392e-03 0.2501498 0.318 0.219 1.000000e+00 1 ## Taf5 3.878714e-03 0.2293076 0.237 0.154 1.000000e+00 1 ## Syt3 3.887045e-03 1.4161966 0.037 0.012 1.000000e+00 1 ## Nupr2 3.897133e-03 0.9420429 0.082 0.039 1.000000e+00 1 ## Sfxn4 3.909815e-03 0.3697310 0.078 0.036 1.000000e+00 1 ## Tsga10 3.912810e-03 0.3639530 0.114 0.061 1.000000e+00 1 ## Mbtps2 3.926498e-03 0.1287151 0.241 0.154 1.000000e+00 1 ## Eif3c1 3.946789e-03 0.1097274 0.976 0.792 1.000000e+00 1 ## Rassf6 3.958212e-03 0.2772255 0.257 0.171 1.000000e+00 1 ## Gm11337 3.982179e-03 2.9319981 0.012 0.001 1.000000e+00 1 ## LOC118568635 4.001321e-03 3.6741074 0.012 0.001 1.000000e+00 1 ## Gm33149 4.001321e-03 2.9764429 0.012 0.001 1.000000e+00 1 ## Slc4a1ap 4.002091e-03 0.1043101 0.314 0.215 1.000000e+00 1 ## Ccdc62 4.009728e-03 0.7266439 0.094 0.048 1.000000e+00 1 ## Larp7 4.012403e-03 0.1271974 0.400 0.280 1.000000e+00 1 ## Lrrc61 4.013464e-03 0.5744989 0.151 0.089 1.000000e+00 1 ## B3gnt7 4.017287e-03 0.2303952 0.143 0.081 1.000000e+00 1 ## Lrtm2 4.020546e-03 3.0340784 0.012 0.001 1.000000e+00 1 ## Gm53375 4.020546e-03 3.0656279 0.012 0.001 1.000000e+00 1 ## E130310I04Rik 4.020546e-03 2.9301667 0.012 0.001 1.000000e+00 1 ## 9330151L19Rik 4.028015e-03 0.8484689 0.106 0.057 1.000000e+00 1 ## D030040B21Rik 4.039855e-03 2.6790497 0.012 0.001 1.000000e+00 1 ## 4930487H11Rik 4.039855e-03 2.8632479 0.012 0.001 1.000000e+00 1 ## Gm19708 4.039855e-03 2.9854172 0.012 0.001 1.000000e+00 1 ## Gm32592 4.039855e-03 2.8853357 0.012 0.001 1.000000e+00 1 ## Gm41406 4.039855e-03 2.7714834 0.012 0.001 1.000000e+00 1 ## Actn3 4.039855e-03 2.9424425 0.012 0.001 1.000000e+00 1 ## 9430060I03Rik 4.053834e-03 0.7719320 0.069 0.031 1.000000e+00 1 ## Gm50627 4.059248e-03 1.9088025 0.012 0.001 1.000000e+00 1 ## Gm39188 4.059248e-03 2.7629469 0.012 0.001 1.000000e+00 1 ## Gm39887 4.078725e-03 2.6738531 0.012 0.001 1.000000e+00 1 ## Gm47792 4.078725e-03 2.5662070 0.012 0.001 1.000000e+00 1 ## Gm57794 4.098288e-03 1.5662634 0.012 0.001 1.000000e+00 1 ## Gm46174 4.098288e-03 2.3956206 0.012 0.001 1.000000e+00 1 ## Gm53300 4.098288e-03 2.4888457 0.012 0.001 1.000000e+00 1 ## Gm16244 4.098288e-03 2.5781396 0.012 0.001 1.000000e+00 1 ## Gm30025 4.121905e-03 0.7951307 0.167 0.104 1.000000e+00 1 ## Gm46137 4.134579e-03 1.1024512 0.049 0.019 1.000000e+00 1 ## Adat1 4.151948e-03 0.2655701 0.143 0.083 1.000000e+00 1 ## Hdac11 4.157105e-03 0.1584437 0.294 0.200 1.000000e+00 1 ## Yy2 4.161967e-03 0.7347590 0.073 0.034 1.000000e+00 1 ## Alg31 4.168709e-03 0.2578859 0.241 0.158 1.000000e+00 1 ## Zfp316 4.186962e-03 0.5940921 0.131 0.075 1.000000e+00 1 ## 5830444B04Rik 4.220323e-03 0.1785506 0.208 0.129 1.000000e+00 1 ## Irgq 4.280616e-03 0.1960074 0.371 0.261 1.000000e+00 1 ## Zbtb39 4.355589e-03 0.4543985 0.159 0.096 1.000000e+00 1 ## Fbxl9 4.421423e-03 0.6787916 0.069 0.031 1.000000e+00 1 ## 1810062G17Rik 4.439659e-03 0.7542847 0.069 0.031 1.000000e+00 1 ## Glb1l 4.468357e-03 0.4902718 0.090 0.045 1.000000e+00 1 ## Vcpkmt 4.469888e-03 0.4720302 0.180 0.115 1.000000e+00 1 ## Cep20 4.473531e-03 0.1312202 0.494 0.358 1.000000e+00 1 ## Usp46 4.513750e-03 0.2170451 0.273 0.182 1.000000e+00 1 ## Gm53985 4.539659e-03 2.4058580 0.016 0.003 1.000000e+00 1 ## Gm46901 4.576818e-03 0.4131213 0.065 0.029 1.000000e+00 1 ## 9130019P16Rik 4.590981e-03 2.4295008 0.016 0.003 1.000000e+00 1 ## Gm39424 4.590981e-03 1.9200310 0.016 0.003 1.000000e+00 1 ## Ssc4d 4.596067e-03 0.6030012 0.082 0.039 1.000000e+00 1 ## Nudt6 4.623683e-03 0.8060851 0.094 0.049 1.000000e+00 1 ## Gm30365 4.625487e-03 1.8287594 0.016 0.003 1.000000e+00 1 ## Zfp946 4.629054e-03 0.1856050 0.371 0.262 1.000000e+00 1 ## Gm30934 4.660228e-03 2.1196453 0.016 0.003 1.000000e+00 1 ## Bloc1s4 4.663399e-03 0.2698120 0.204 0.131 1.000000e+00 1 ## Armcx5 4.683744e-03 0.2023585 0.204 0.130 1.000000e+00 1 ## Rfc3 4.698731e-03 0.2373286 0.216 0.140 1.000000e+00 1 ## Lig4 4.703080e-03 0.2103007 0.257 0.170 1.000000e+00 1 ## Bbs10 4.734458e-03 0.7568675 0.057 0.024 1.000000e+00 1 ## Pex141 4.748019e-03 0.2848240 0.314 0.219 1.000000e+00 1 ## Wdr18 4.758293e-03 0.1619485 0.318 0.211 1.000000e+00 1 ## Rexo5 4.806667e-03 0.5037239 0.086 0.043 1.000000e+00 1 ## Ccdc166 4.820251e-03 0.4092402 0.180 0.110 1.000000e+00 1 ## Zfp97 4.820438e-03 0.6774742 0.086 0.043 1.000000e+00 1 ## Zfp108 4.830151e-03 1.0545058 0.061 0.027 1.000000e+00 1 ## Zfp27 4.859631e-03 0.4108392 0.102 0.054 1.000000e+00 1 ## Sh2d3c 4.911206e-03 0.2850292 0.204 0.127 1.000000e+00 1 ## Cdk17 4.968332e-03 0.1218039 0.180 0.110 1.000000e+00 1 ## Nts 4.978155e-03 0.2177264 0.967 0.905 1.000000e+00 1 ## Telo2 4.979594e-03 0.4483570 0.131 0.075 1.000000e+00 1 ## Mt3 4.991114e-03 0.1294197 0.131 0.073 1.000000e+00 1 ## Arl14ep 5.085230e-03 0.2710597 0.204 0.131 1.000000e+00 1 ## Zfp958 5.181920e-03 0.3948515 0.180 0.112 1.000000e+00 1 ## Mre11a 5.201937e-03 0.1993570 0.216 0.140 1.000000e+00 1 ## Abcb4 5.209697e-03 1.0346659 0.041 0.015 1.000000e+00 1 ## Zfp984 5.230163e-03 0.3823757 0.192 0.121 1.000000e+00 1 ## Cmss1 5.257917e-03 0.9562362 0.102 0.056 1.000000e+00 1 ## Elk1 5.273195e-03 0.7643759 0.098 0.052 1.000000e+00 1 ## 3110045C21Rik 5.313580e-03 0.8357508 0.057 0.024 1.000000e+00 1 ## Gm32856 5.353818e-03 0.1630869 0.131 0.074 1.000000e+00 1 ## Insl5 5.381738e-03 0.1279706 0.024 0.006 1.000000e+00 1 ## Arsk 5.390025e-03 0.5230100 0.102 0.055 1.000000e+00 1 ## Matn2 5.426981e-03 2.2550710 0.033 0.010 1.000000e+00 1 ## Elovl2 5.430814e-03 0.5281864 0.078 0.037 1.000000e+00 1 ## Setd4 5.458177e-03 0.4768848 0.086 0.043 1.000000e+00 1 ## Kif9 5.467985e-03 0.6285581 0.065 0.029 1.000000e+00 1 ## Gm41466 5.469547e-03 1.6174592 0.024 0.006 1.000000e+00 1 ## Gm54057 5.499103e-03 1.3084772 0.024 0.006 1.000000e+00 1 ## Cyp39a1 5.508290e-03 0.1857710 0.082 0.041 1.000000e+00 1 ## Pdxp 5.571323e-03 2.3601511 0.029 0.008 1.000000e+00 1 ## Trpc3 5.585475e-03 1.2399842 0.037 0.013 1.000000e+00 1 ## Tmlhe 5.658503e-03 0.8448373 0.090 0.047 1.000000e+00 1 ## Rbm12b1 5.753337e-03 0.1813651 0.163 0.099 1.000000e+00 1 ## Mrpl49 5.760120e-03 0.1046159 0.408 0.284 1.000000e+00 1 ## C030034I22Rik 5.766182e-03 0.1968070 0.171 0.106 1.000000e+00 1 ## Tubg2 5.766282e-03 0.4231249 0.078 0.037 1.000000e+00 1 ## A930006K02Rik 5.850368e-03 0.9416253 0.057 0.024 1.000000e+00 1 ## Ankrd49 5.852224e-03 0.1571472 0.282 0.190 1.000000e+00 1 ## Zfp719 5.860016e-03 0.1014097 0.220 0.143 1.000000e+00 1 ## Gm36684 5.871745e-03 1.4170308 0.029 0.008 1.000000e+00 1 ## Xrcc2 5.913824e-03 1.7879280 0.029 0.008 1.000000e+00 1 ## Efnb2 5.917050e-03 0.1103258 0.253 0.166 1.000000e+00 1 ## Ints14 5.919949e-03 0.1205196 0.229 0.149 1.000000e+00 1 ## Gm10244 5.927910e-03 1.6807875 0.029 0.008 1.000000e+00 1 ## Zfp84 5.942881e-03 0.1276268 0.347 0.242 1.000000e+00 1 ## Styx 6.006934e-03 0.2255036 0.249 0.171 1.000000e+00 1 ## Rtl8c 6.013066e-03 1.6067350 0.029 0.008 1.000000e+00 1 ## Cngb1 6.041696e-03 2.0932900 0.029 0.008 1.000000e+00 1 ## Gm46043 6.088850e-03 1.6450760 0.033 0.010 1.000000e+00 1 ## Pde8b 6.130927e-03 0.1545104 0.176 0.109 1.000000e+00 1 ## AU041133 6.131426e-03 0.1434476 0.306 0.211 1.000000e+00 1 ## Sv2a 6.132354e-03 0.6426472 0.078 0.038 1.000000e+00 1 ## Zfp433 6.145832e-03 0.4944569 0.057 0.024 1.000000e+00 1 ## Lrrc40 6.212905e-03 0.3710974 0.147 0.089 1.000000e+00 1 ## Tnfsf13b 6.214410e-03 1.0323471 0.053 0.022 1.000000e+00 1 ## Crtc1 6.217771e-03 0.1705551 0.273 0.181 1.000000e+00 1 ## Hdgfl2 6.236598e-03 0.2149703 0.363 0.261 1.000000e+00 1 ## Gm13524 6.248112e-03 1.0150077 0.065 0.030 1.000000e+00 1 ## Iqcg 6.258729e-03 0.4996341 0.098 0.052 1.000000e+00 1 ## Rab26os 6.266812e-03 0.6956254 0.127 0.075 1.000000e+00 1 ## Trim5 6.267221e-03 0.5555612 0.061 0.027 1.000000e+00 1 ## Gm51817 6.275415e-03 1.4698217 0.033 0.010 1.000000e+00 1 ## Slc10a3 6.278618e-03 0.3325253 0.110 0.061 1.000000e+00 1 ## Gm34045 6.310217e-03 0.4316048 0.037 0.013 1.000000e+00 1 ## Gm40208 6.364432e-03 1.3320112 0.045 0.017 1.000000e+00 1 ## Top1mt 6.388166e-03 0.7393457 0.086 0.045 1.000000e+00 1 ## Clasp2 6.398044e-03 0.1565194 0.241 0.161 1.000000e+00 1 ## Lmo2 6.419532e-03 0.9829715 0.045 0.017 1.000000e+00 1 ## Wrap53 6.447588e-03 0.1869131 0.188 0.120 1.000000e+00 1 ## Fkrp1 6.448011e-03 0.1239183 0.355 0.242 1.000000e+00 1 ## Gm14325 6.463969e-03 0.5051738 0.147 0.089 1.000000e+00 1 ## Cdyl2 6.489550e-03 1.0472188 0.053 0.022 1.000000e+00 1 ## Prmt9 6.594071e-03 0.2482556 0.208 0.134 1.000000e+00 1 ## Klf16 6.677760e-03 0.1090536 0.200 0.128 1.000000e+00 1 ## Rwdd3 6.727781e-03 1.0751487 0.057 0.025 1.000000e+00 1 ## Ap3m2 6.746334e-03 0.1224161 0.180 0.111 1.000000e+00 1 ## Gm31364 6.868665e-03 0.6718373 0.065 0.030 1.000000e+00 1 ## Poc5 6.870926e-03 0.1685127 0.184 0.116 1.000000e+00 1 ## Zfp354b 6.902770e-03 0.5612608 0.090 0.047 1.000000e+00 1 ## Bckdhb 6.926431e-03 0.9353157 0.073 0.036 1.000000e+00 1 ## Xab2 6.975886e-03 0.2361561 0.302 0.212 1.000000e+00 1 ## Gm10463 7.016547e-03 1.1775453 0.041 0.015 1.000000e+00 1 ## Nop9 7.021260e-03 0.1296437 0.273 0.184 1.000000e+00 1 ## Gm38673 7.056863e-03 0.7345180 0.086 0.044 1.000000e+00 1 ## Acp6 7.103444e-03 0.1544175 0.302 0.210 1.000000e+00 1 ## Ubxn2b 7.148619e-03 0.2853630 0.171 0.106 1.000000e+00 1 ## Slc26a11 7.209879e-03 0.7753631 0.049 0.020 1.000000e+00 1 ## Adamts15 7.239029e-03 0.1872741 0.114 0.064 1.000000e+00 1 ## Gm51801 7.276232e-03 0.6373861 0.078 0.038 1.000000e+00 1 ## Adrb1 7.314050e-03 1.3232539 0.041 0.015 1.000000e+00 1 ## 6330418K02Rik 7.322963e-03 0.3111788 0.139 0.082 1.000000e+00 1 ## Scfd2 7.387264e-03 0.2963181 0.135 0.080 1.000000e+00 1 ## Mocs3 7.406847e-03 0.4886067 0.118 0.068 1.000000e+00 1 ## Tusc1 7.418988e-03 0.5700141 0.147 0.090 1.000000e+00 1 ## Wdr91 7.431837e-03 0.9663736 0.049 0.020 1.000000e+00 1 ## Gm36117 7.431837e-03 1.0546260 0.049 0.020 1.000000e+00 1 ## Riox1 7.497955e-03 0.3059465 0.184 0.121 1.000000e+00 1 ## Faxc 7.665730e-03 0.2061013 0.127 0.072 1.000000e+00 1 ## Gm50602 7.675491e-03 0.6994961 0.065 0.031 1.000000e+00 1 ## Gm15483 7.684272e-03 1.0678585 0.049 0.020 1.000000e+00 1 ## Oxnad1 7.689068e-03 0.4010567 0.192 0.124 1.000000e+00 1 ## Numbl 7.705270e-03 1.0680835 0.041 0.015 1.000000e+00 1 ## Rpl37rt 7.777621e-03 0.1269911 0.302 0.207 1.000000e+00 1 ## BC037039 7.838057e-03 2.0445865 0.020 0.005 1.000000e+00 1 ## Tmem65 7.910708e-03 0.1224074 0.155 0.096 1.000000e+00 1 ## Cgnl1 7.928920e-03 1.4420970 0.061 0.029 1.000000e+00 1 ## Dynlt1b 7.992651e-03 0.9640502 0.061 0.028 1.000000e+00 1 ## Abraxas1 8.000380e-03 0.5904829 0.110 0.064 1.000000e+00 1 ## Rgs12 8.054958e-03 0.9508638 0.106 0.061 1.000000e+00 1 ## Mcub 8.134946e-03 0.1722642 0.049 0.020 1.000000e+00 1 ## Meak7 8.156290e-03 0.3348101 0.114 0.064 1.000000e+00 1 ## Pcdhb15 8.162542e-03 1.6282886 0.020 0.005 1.000000e+00 1 ## Spin4 8.204266e-03 1.5760570 0.037 0.013 1.000000e+00 1 ## Slc35b4 8.341106e-03 0.3969522 0.167 0.106 1.000000e+00 1 ## Gm36359 8.382216e-03 0.8479635 0.049 0.020 1.000000e+00 1 ## Stam 8.435246e-03 0.1251594 0.335 0.239 1.000000e+00 1 ## Gm40733 8.505414e-03 1.1355184 0.037 0.013 1.000000e+00 1 ## Gm14393 8.510511e-03 0.4865046 0.073 0.036 1.000000e+00 1 ## Gm32313 8.537682e-03 0.8952581 0.037 0.013 1.000000e+00 1 ## Hykk 8.547916e-03 0.7046806 0.065 0.031 1.000000e+00 1 ## Gm38486 8.553857e-03 1.0528272 0.037 0.013 1.000000e+00 1 ## Spire2 8.563083e-03 0.1898505 0.253 0.174 1.000000e+00 1 ## 1700018L02Rik 8.570059e-03 1.4158690 0.037 0.013 1.000000e+00 1 ## Asb1 8.579572e-03 0.4611719 0.139 0.083 1.000000e+00 1 ## Gm36449 8.653410e-03 0.2835510 0.237 0.161 1.000000e+00 1 ## Hspa13 8.666217e-03 0.1425398 0.461 0.334 1.000000e+00 1 ## Rpgrip1 8.705837e-03 0.1818642 0.180 0.115 1.000000e+00 1 ## Gm4285 8.747049e-03 0.3693431 0.151 0.093 1.000000e+00 1 ## Casp12 8.764693e-03 1.7364548 0.033 0.011 1.000000e+00 1 ## Cyb561d2 8.818328e-03 0.2998805 0.322 0.231 1.000000e+00 1 ## Alg1 8.856653e-03 0.2289002 0.216 0.143 1.000000e+00 1 ## Gm36856 8.941656e-03 1.0429969 0.057 0.026 1.000000e+00 1 ## B4galt71 8.969359e-03 0.1802732 0.216 0.140 1.000000e+00 1 ## Stamos 9.058561e-03 0.3570167 0.110 0.063 1.000000e+00 1 ## Pigb 9.067128e-03 0.2557653 0.110 0.063 1.000000e+00 1 ## Gm36447 9.072861e-03 1.3816180 0.033 0.011 1.000000e+00 1 ## Gm54309 9.110527e-03 0.6788258 0.045 0.018 1.000000e+00 1 ## 9030624G23Rik 9.144751e-03 0.8318399 0.057 0.026 1.000000e+00 1 ## 4930511A08Rik 9.155616e-03 2.3545220 0.024 0.007 1.000000e+00 1 ## Polr3f 9.203405e-03 0.1104868 0.180 0.114 1.000000e+00 1 ## Gm13572 9.223866e-03 2.2670191 0.024 0.007 1.000000e+00 1 ## Gm30074 9.263359e-03 0.6740406 0.078 0.039 1.000000e+00 1 ## Banf2 9.292570e-03 1.8687618 0.024 0.007 1.000000e+00 1 ## Gm53451 9.337088e-03 0.5086004 0.045 0.018 1.000000e+00 1 ## Cetn4 9.413514e-03 0.9933495 0.065 0.031 1.000000e+00 1 ## Oas1c 9.442576e-03 0.2815554 0.053 0.023 1.000000e+00 1 ## Gm54038 9.478018e-03 1.5452220 0.024 0.007 1.000000e+00 1 ## Erc2 9.568663e-03 0.9064219 0.045 0.018 1.000000e+00 1 ## Gm40371 9.619263e-03 0.8515805 0.024 0.007 1.000000e+00 1 ## Rab8b 9.633768e-03 0.4598548 0.143 0.089 1.000000e+00 1 ## Gm46933 9.697598e-03 0.6402879 0.057 0.026 1.000000e+00 1 ## Zkscan4 9.719761e-03 1.5037634 0.029 0.009 1.000000e+00 1 ## Cab39l 9.769574e-03 0.1045877 0.306 0.216 1.000000e+00 1 ## Wdr55 9.780295e-03 0.1388795 0.167 0.107 1.000000e+00 1 ## Mmrn2 9.784203e-03 1.2628342 0.029 0.009 1.000000e+00 1 ## Wdr35 9.863182e-03 0.3672824 0.135 0.080 1.000000e+00 1 ## Gm38604 9.870718e-03 1.5638803 0.029 0.009 1.000000e+00 1 ## Shroom4 9.870718e-03 1.4062964 0.029 0.009 1.000000e+00 1 ## Gm40572 9.883093e-03 1.2571076 0.024 0.007 1.000000e+00 1 ## Flrt3 9.915094e-03 0.6731260 0.122 0.075 1.000000e+00 1 ## Mtg2 9.918155e-03 0.1303947 0.261 0.179 1.000000e+00 1 ## Gm34362 9.936052e-03 0.9101218 0.029 0.009 1.000000e+00 1 ## Asb3 9.961002e-03 0.6505399 0.139 0.087 1.000000e+00 1 ## Lrrc28 9.997782e-03 0.4371728 0.135 0.082 1.000000e+00 1 ## gene ## Pzp Pzp ## Habp2 Habp2 ## Crp Crp ## Adamts5 Adamts5 ## Cyp2j5 Cyp2j5 ## Ugt8a Ugt8a ## Basp1 Basp1 ## Mc4r Mc4r ## Col17a1 Col17a1 ## Kcnk16 Kcnk16 ## Galnt13 Galnt13 ## Tlr2 Tlr2 ## Casr Casr ## Rbm24 Rbm24 ## Gm33649 Gm33649 ## Onecut3 Onecut3 ## Crtac1 Crtac1 ## Tlr5 Tlr5 ## Cltrn Cltrn ## Smbd1 Smbd1 ## Zfp618 Zfp618 ## Pgc Pgc ## E030025P04Rik E030025P04Rik ## Cyp2j6 Cyp2j6 ## Tril Tril ## Agr2 Agr2 ## Gm53467 Gm53467 ## Agr31 Agr3 ## D930028M14Rik D930028M14Rik ## Slc39a8 Slc39a8 ## Cck Cck ## Scg2 Scg2 ## Scn9a Scn9a ## Gm12511 Gm12511 ## Etv1 Etv1 ## Upp1 Upp1 ## Rflna Rflna ## Parm1 Parm1 ## Scn3b Scn3b ## U90926 U90926 ## Cpn1 Cpn1 ## Cux2 Cux2 ## Cst3 Cst3 ## Tle1 Tle1 ## Fmo5 Fmo5 ## 3110004A20Rik 3110004A20Rik ## Tmem176a Tmem176a ## Gm11992 Gm11992 ## Proc Proc ## Egfl6 Egfl6 ## Spart Spart ## Vkorc1 Vkorc1 ## Arx Arx ## Rdh7 Rdh7 ## Lyzl4 Lyzl4 ## Tec Tec ## Tff31 Tff3 ## Slc41a2 Slc41a2 ## Dclk3 Dclk3 ## S100a11 S100a11 ## Cit Cit ## Rfxap Rfxap ## Cav2 Cav2 ## Aldoc Aldoc ## Ms4a10 Ms4a10 ## Homer21 Homer2 ## Fgl2 Fgl2 ## Slc8a1 Slc8a1 ## Pla2g12a Pla2g12a ## Fars2 Fars2 ## Rbp4 Rbp4 ## Igfbp7 Igfbp7 ## Oxr1 Oxr1 ## Itm2b Itm2b ## Slc25a4 Slc25a4 ## Tm4sf41 Tm4sf4 ## Ms4a8a1 Ms4a8a ## Dapp1 Dapp1 ## Nenf Nenf ## Aig1 Aig1 ## Ly6e Ly6e ## Prox1 Prox1 ## Ifitm2 Ifitm2 ## Bace2 Bace2 ## Marveld1 Marveld1 ## Grpr Grpr ## Me3 Me3 ## Gm27162 Gm27162 ## Tmem176b Tmem176b ## Trpm3 Trpm3 ## Cyp2j9 Cyp2j9 ## Prss23 Prss23 ## Lgals21 Lgals2 ## Qpct Qpct ## Gm46558 Gm46558 ## Sting1 Sting1 ## Ugt2b34 Ugt2b34 ## Myl7 Myl7 ## Dner Dner ## Tspan12 Tspan12 ## Casp6 Casp6 ## Plac81 Plac8 ## Klkb11 Klkb1 ## Gm46905 Gm46905 ## Serpina1b1 Serpina1b ## Wars1 Wars1 ## Tshz2 Tshz2 ## Abca8b Abca8b ## Krt201 Krt20 ## Atp8a2 Atp8a2 ## Sstr2 Sstr2 ## Gm17619 Gm17619 ## Axin2 Axin2 ## Tmed6 Tmed6 ## Tpst2 Tpst2 ## Serpine2 Serpine2 ## Anxa13 Anxa13 ## Apom Apom ## Dio1 Dio1 ## Upk1b Upk1b ## Sct Sct ## Egfl7 Egfl7 ## Psap Psap ## Gclm Gclm ## Gm52807 Gm52807 ## Smarca1 Smarca1 ## Cryba4 Cryba4 ## Rflnb Rflnb ## Shf Shf ## Gast Gast ## Bsg1 Bsg ## Ano6 Ano6 ## Spry1 Spry1 ## F51 F5 ## Ndrg11 Ndrg1 ## Aga1 Aga ## Lrpap1 Lrpap1 ## Trf1 Trf ## Tmed3 Tmed3 ## Uts2b Uts2b ## Kcnj6 Kcnj6 ## Cops6 Cops6 ## Cpm Cpm ## Cpe Cpe ## Ssr4 Ssr4 ## Rps20 Rps20 ## Col6a5 Col6a5 ## Tmem254 Tmem254 ## Gnai1 Gnai1 ## Atoh8 Atoh8 ## Rps24 Rps24 ## Txndc5 Txndc5 ## Cyp2c55 Cyp2c55 ## Gm53234 Gm53234 ## Smad9 Smad9 ## Rpl32 Rpl32 ## Ppic1 Ppic ## Bambi Bambi ## Rasgrf2 Rasgrf2 ## 4732490B19Rik 4732490B19Rik ## Gm9767 Gm9767 ## Mtcl2 Mtcl2 ## Ihh Ihh ## Rps27a Rps27a ## Rps18 Rps18 ## Pcbd11 Pcbd1 ## Pabpc1l Pabpc1l ## Exoc7 Exoc7 ## Gcnt3 Gcnt3 ## Hunk1 Hunk ## Cited2 Cited2 ## Gnas Gnas ## Fxyd31 Fxyd3 ## Gimd11 Gimd1 ## Pxylp1 Pxylp1 ## Gm30524 Gm30524 ## 4930573H18Rik 4930573H18Rik ## Mill2 Mill2 ## Abcb1a Abcb1a ## Gfra1 Gfra1 ## Gm2102 Gm2102 ## Anxa2 Anxa2 ## Rpl38 Rpl38 ## Fut2 Fut2 ## Apcs Apcs ## Ghrl Ghrl ## Cfi Cfi ## Cldn16 Cldn16 ## Sis1 Sis ## Pde3a Pde3a ## Efcab151 Efcab15 ## C2 C2 ## Camk4 Camk4 ## Plgrkt Plgrkt ## Gm30986 Gm30986 ## Cyp4f14 Cyp4f14 ## Nfix Nfix ## Spint21 Spint2 ## Nefl Nefl ## Trappc9 Trappc9 ## Sema4g Sema4g ## Pnma8b Pnma8b ## Cdkl1 Cdkl1 ## 2610524H06Rik 2610524H06Rik ## Slc12a8 Slc12a8 ## Nbdy Nbdy ## Rpl19 Rpl19 ## Atp5mk1 Atp5mk ## H2-D1 H2-D1 ## Rtn4r Rtn4r ## Gabarapl21 Gabarapl2 ## Rps3 Rps3 ## Pfkfb3 Pfkfb3 ## Tspo Tspo ## Rps10 Rps10 ## Snhg18 Snhg18 ## Ebpl Ebpl ## Nostrin1 Nostrin ## Pir Pir ## Lgals3bp Lgals3bp ## Tmod2 Tmod2 ## Tpst11 Tpst1 ## Rps2 Rps2 ## Rpl39 Rpl39 ## Olfm11 Olfm1 ## Cr1l Cr1l ## Fads2 Fads2 ## Pcbd2 Pcbd2 ## Efcc1 Efcc1 ## Krt79 Krt79 ## Ldb3 Ldb3 ## Gm39858 Gm39858 ## Rpl12 Rpl12 ## Cyp2c681 Cyp2c68 ## Col11a2 Col11a2 ## Hnf1aos1 Hnf1aos1 ## Cldn1 Cldn1 ## Slit1 Slit1 ## Mycn Mycn ## Ccdc85b Ccdc85b ## Tm7sf2 Tm7sf2 ## Pepd Pepd ## 5033421B08Rik 5033421B08Rik ## Maged2 Maged2 ## Mgll1 Mgll ## Cnpy2 Cnpy2 ## Slc16a10 Slc16a10 ## Bhlhe41 Bhlhe41 ## Lysmd21 Lysmd2 ## Zfp395 Zfp395 ## Pglyrp1 Pglyrp1 ## Amotl2 Amotl2 ## Hgfac Hgfac ## Endod1 Endod1 ## B3gnt9 B3gnt9 ## Ctdspl Ctdspl ## Elapor11 Elapor1 ## Sh3pxd2a Sh3pxd2a ## Rpl15 Rpl15 ## Ywhah Ywhah ## Nog Nog ## Rpl37a1 Rpl37a ## Tpmt Tpmt ## Ttr1 Ttr ## Cap2 Cap2 ## Calml41 Calml4 ## Rps7 Rps7 ## Uchl1 Uchl1 ## Tdrd12 Tdrd12 ## Arsb1 Arsb ## H2-K1 H2-K1 ## Hexb Hexb ## Fam222a Fam222a ## Ppp1r14a Ppp1r14a ## Snx18 Snx18 ## Ypel3 Ypel3 ## Qsox1 Qsox1 ## Cpq Cpq ## Ptp4a3 Ptp4a3 ## Fam174a1 Fam174a ## Muc4 Muc4 ## Prdx11 Prdx1 ## Gm30648 Gm30648 ## Rpl37 Rpl37 ## Rpl27a Rpl27a ## Nkx6-3 Nkx6-3 ## Klf7 Klf7 ## Epha4 Epha4 ## Kcnj3 Kcnj3 ## Ctsf Ctsf ## Ggact Ggact ## Ces2a Ces2a ## Mrpl15 Mrpl15 ## Vsig2 Vsig2 ## Rnf130 Rnf130 ## Blnk Blnk ## Riiad11 Riiad1 ## Acaa2 Acaa2 ## Tomm7 Tomm7 ## Atp5mc2 Atp5mc2 ## Svip1 Svip ## Fxyd5 Fxyd5 ## 9330162012Rik 9330162012Rik ## Rpl13 Rpl13 ## Rpl3 Rpl3 ## Rps4x Rps4x ## Hagh Hagh ## Rap1gap Rap1gap ## Mpc2 Mpc2 ## Usp25 Usp25 ## Klf12 Klf12 ## Tspan11 Tspan1 ## Commd1 Commd1 ## Cped1 Cped1 ## Ifi271 Ifi27 ## Cfap69 Cfap69 ## Tulp2 Tulp2 ## Bcl11a Bcl11a ## Kif2a Kif2a ## Trp53inp1 Trp53inp1 ## Slc50a1 Slc50a1 ## Mettl261 Mettl26 ## Pdzk1ip1 Pdzk1ip1 ## 4732419C18Rik 4732419C18Rik ## Rps3a1 Rps3a1 ## Rpl23 Rpl23 ## Gm39460 Gm39460 ## Bcam1 Bcam ## Rps16 Rps16 ## Rps15a Rps15a ## Mia Mia ## Atp5if1 Atp5if1 ## Rnaset2a Rnaset2a ## Grcc101 Grcc10 ## Tcaf2 Tcaf2 ## Tsc22d11 Tsc22d1 ## Urah Urah ## Shisal1 Shisal1 ## Msrb1 Msrb1 ## Marchf11 Marchf11 ## Mtcl1 Mtcl1 ## Ppia1 Ppia ## Cga Cga ## Grb14 Grb14 ## Rps13 Rps13 ## Pla2g2d Pla2g2d ## Gcg Gcg ## Scgn Scgn ## Gde1 Gde1 ## Gm41848 Gm41848 ## Gm142071 Gm14207 ## Rpl11 Rpl11 ## Slc22a15 Slc22a15 ## Dip2a Dip2a ## Ikbip Ikbip ## Lsm6 Lsm6 ## Trim2 Trim2 ## Romo11 Romo1 ## Rpl17 Rpl17 ## Ceacam10 Ceacam10 ## Glod51 Glod5 ## Cdkl5 Cdkl5 ## Agfg2 Agfg2 ## Tmem263 Tmem263 ## Lin7a Lin7a ## G430095P16Rik G430095P16Rik ## Ret1 Ret ## Eef1a1 Eef1a1 ## Rac3 Rac3 ## Nhlh1 Nhlh1 ## Tbc1d9 Tbc1d9 ## Rpl34 Rpl34 ## Leprotl11 Leprotl1 ## Sh3bgrl3 Sh3bgrl3 ## Gm53846 Gm53846 ## Lbp Lbp ## Cd59a Cd59a ## Rnaset2b Rnaset2b ## Ero1b Ero1b ## P2rx4 P2rx4 ## Diras2 Diras2 ## Sgpp2 Sgpp2 ## F13b F13b ## Mgst2 Mgst2 ## Rpl35a Rpl35a ## Uba52 Uba52 ## Rpl22l1 Rpl22l1 ## Kcnh31 Kcnh3 ## Rpl18 Rpl18 ## Stmn3 Stmn3 ## Rprm Rprm ## Hint1 Hint1 ## Ndufa1 Ndufa1 ## Lyplal1 Lyplal1 ## Fth1 Fth1 ## Rab37 Rab37 ## Ptms Ptms ## Hus1 Hus1 ## Il1rap Il1rap ## Arl10 Arl10 ## Susd4 Susd4 ## Gadd45b1 Gadd45b ## C8b C8b ## Pgap1 Pgap1 ## Pex19 Pex19 ## Chchd10 Chchd10 ## Rgs17 Rgs17 ## B4galt4 B4galt4 ## Serpina1d1 Serpina1d ## Orm3 Orm3 ## Znrf2 Znrf2 ## Prdx5 Prdx5 ## Mblac2 Mblac2 ## Cmas1 Cmas ## Anp32a Anp32a ## Sgpl11 Sgpl1 ## Gm53258 Gm53258 ## LOC118568133 LOC118568133 ## Lst1 Lst1 ## H3f3a1 H3f3a ## 1810037I17Rik 1810037I17Rik ## Pkp1 Pkp1 ## C4bp C4bp ## Pcca1 Pcca ## Slc17a4 Slc17a4 ## Mien11 Mien1 ## Txnip Txnip ## Mfsd2a Mfsd2a ## Pbld2 Pbld2 ## Pebp1 Pebp1 ## Isg20 Isg20 ## Cyb5r31 Cyb5r3 ## Rplp0 Rplp0 ## Gstm1 Gstm1 ## Greb1 Greb1 ## Scg51 Scg5 ## Sdc2 Sdc2 ## Gpld1 Gpld1 ## Dgkk Dgkk ## A2ml1 A2ml1 ## Lims1 Lims1 ## Ndufb12 Ndufb1 ## Rpl6 Rpl6 ## Ndufb11 Ndufb11 ## Gprc5c Gprc5c ## Tmem134 Tmem134 ## Rasa4 Rasa4 ## Hcfc1r1 Hcfc1r1 ## Efcab12 Efcab12 ## Dap Dap ## Akr1b11 Akr1b1 ## Plpp3 Plpp3 ## Fhit Fhit ## Pter Pter ## Plekhg1 Plekhg1 ## Rpl29 Rpl29 ## Rps14 Rps14 ## Rpl7 Rpl7 ## Ifnar2 Ifnar2 ## Man2b21 Man2b2 ## Pparg Pparg ## Gm34173 Gm34173 ## Soat1 Soat1 ## Gm5617 Gm5617 ## Galnt7 Galnt7 ## Tfpi Tfpi ## Enpp3 Enpp3 ## Tcaf1 Tcaf1 ## Adgrf5 Adgrf5 ## Stard4 Stard4 ## Cyb5a Cyb5a ## Rexo2 Rexo2 ## Rgs21 Rgs2 ## Lypla11 Lypla1 ## 1110065P20Rik 1110065P20Rik ## Focad Focad ## Naglu Naglu ## Eif1b Eif1b ## Ctsb Ctsb ## Slc48a1 Slc48a1 ## Tmem38b Tmem38b ## Cbr1 Cbr1 ## Asl Asl ## Cdkn1c1 Cdkn1c ## Mrpl331 Mrpl33 ## Parva Parva ## LOC118567825 LOC118567825 ## Stx17 Stx17 ## Frrs1 Frrs1 ## Tmem97 Tmem97 ## Rcan3 Rcan3 ## Ctnnb11 Ctnnb1 ## Ffar4 Ffar4 ## Coa3 Coa3 ## Tomm20 Tomm20 ## Tmem200a1 Tmem200a ## 9530068E07Rik1 9530068E07Rik ## Atp6v1f1 Atp6v1f ## Zfyve21 Zfyve21 ## Pfdn5 Pfdn5 ## Tcea3 Tcea3 ## Rps8 Rps8 ## Nucb2 Nucb2 ## Slc3a11 Slc3a1 ## Snph Snph ## Eef1b2 Eef1b2 ## Cox7a21 Cox7a2 ## Ndrg21 Ndrg2 ## Sem1 Sem1 ## Sez6l21 Sez6l2 ## Hps4 Hps4 ## Srp141 Srp14 ## Rpl31 Rpl31 ## Hspb6 Hspb6 ## Prom11 Prom1 ## Pid1 Pid1 ## Gstt2 Gstt2 ## Rapgef3 Rapgef3 ## Amer2 Amer2 ## Gas5 Gas5 ## Plekha2 Plekha2 ## Sat2 Sat2 ## Fas Fas ## Ovol2 Ovol2 ## Svil Svil ## Uqcc31 Uqcc3 ## A730098A19Rik A730098A19Rik ## Malrd1 Malrd1 ## Prps2 Prps2 ## Entpd8 Entpd8 ## Tnfaip8l3 Tnfaip8l3 ## Dut Dut ## Gm20234 Gm20234 ## Rps28 Rps28 ## Cfap92 Cfap92 ## Pnlip Pnlip ## Trappc2l Trappc2l ## Rpl30 Rpl30 ## Park7 Park7 ## Slc38a4 Slc38a4 ## C1d C1d ## Tagln3 Tagln3 ## Pmm1 Pmm1 ## Nme2 Nme2 ## Ostc Ostc ## Apobec3 Apobec3 ## Rpl9 Rpl9 ## Tspan17 Tspan17 ## Imp3 Imp3 ## Mcee Mcee ## Psma4 Psma4 ## Rpl8 Rpl8 ## Arl6ip5 Arl6ip5 ## Ebp1 Ebp ## Ccna1 Ccna1 ## Pccb Pccb ## Arl3 Arl3 ## Arpp191 Arpp19 ## Colgalt1 Colgalt1 ## Rab4b Rab4b ## Clec2e Clec2e ## Maob1 Maob ## H2az1 H2az1 ## Kdelr1 Kdelr1 ## Pax6os1 Pax6os1 ## Ogfod3 Ogfod3 ## Strip2 Strip2 ## Sult1d1 Sult1d1 ## Serf2 Serf2 ## Acsl5 Acsl5 ## Tenm3 Tenm3 ## Rps5 Rps5 ## Tet1 Tet1 ## Vps37d Vps37d ## Nfib Nfib ## Prkacb Prkacb ## Arhgap31 Arhgap31 ## Tmem9 Tmem9 ## Galnt41 Galnt4 ## Hopx1 Hopx ## Tm4sf1 Tm4sf1 ## Ocrl Ocrl ## Mmp14 Mmp14 ## Cyp4b1 Cyp4b1 ## Ywhaq1 Ywhaq ## Dnlz Dnlz ## Atxn101 Atxn10 ## Cdc14a Cdc14a ## Pld3 Pld3 ## Gfra3 Gfra3 ## Aplp2 Aplp2 ## Tmx11 Tmx1 ## Mgat31 Mgat3 ## Pgrmc2 Pgrmc2 ## Slc39a2 Slc39a2 ## Mospd21 Mospd2 ## Arhgef38 Arhgef38 ## Psmb9 Psmb9 ## Epha7 Epha7 ## Cela11 Cela1 ## Avil Avil ## Lcmt2 Lcmt2 ## Fads3 Fads3 ## Ezh2 Ezh2 ## Smim6 Smim6 ## Pomgnt2 Pomgnt2 ## Rpl41 Rpl41 ## Rab2a1 Rab2a ## Glo1 Glo1 ## Pla2g2f Pla2g2f ## Sdf41 Sdf4 ## Mexis Mexis ## Ntrk2 Ntrk2 ## Cetn2 Cetn2 ## Pdxk1 Pdxk ## Usp18 Usp18 ## Gm29825 Gm29825 ## Slc16a11 Slc16a11 ## Hepacam21 Hepacam2 ## Zfp4671 Zfp467 ## Maged1 Maged1 ## Id21 Id2 ## Ppib Ppib ## Ergic31 Ergic3 ## Angptl8 Angptl8 ## Col1a2 Col1a2 ## Lrpprc Lrpprc ## Tmod1 Tmod1 ## Npm1 Npm1 ## Cenpx Cenpx ## Pecr Pecr ## Nxnl2 Nxnl2 ## Ost4 Ost4 ## Mapk8ip11 Mapk8ip1 ## Pitx1 Pitx1 ## Slc16a2 Slc16a2 ## Deptor Deptor ## Nit2 Nit2 ## Gm12506 Gm12506 ## Cox171 Cox17 ## Morf4l21 Morf4l2 ## Rps25 Rps25 ## Ap3s11 Ap3s1 ## Golga7b Golga7b ## Bmpr1a Bmpr1a ## Tmem45b Tmem45b ## Cyba Cyba ## Mpc1 Mpc1 ## Rack1 Rack1 ## Tmsb101 Tmsb10 ## Rpl36 Rpl36 ## H2-M3 H2-M3 ## Slc39a14 Slc39a14 ## Gm542261 Gm54226 ## Pfn2 Pfn2 ## Gm34299 Gm34299 ## Rpl14 Rpl14 ## Calm2 Calm2 ## C1qtnf1 C1qtnf1 ## Cfap36 Cfap36 ## Cetn3 Cetn3 ## B2m B2m ## Ffar3 Ffar3 ## Slc40a1 Slc40a1 ## Sv2b Sv2b ## Rheb Rheb ## Nudt2 Nudt2 ## Prkaa21 Prkaa2 ## Dync2li1 Dync2li1 ## Nipsnap3b Nipsnap3b ## Cbfa2t31 Cbfa2t3 ## Rps29 Rps29 ## Bphl Bphl ## Fkbp1a1 Fkbp1a ## Cpd1 Cpd ## Ppp1cb1 Ppp1cb ## Stxbp6 Stxbp6 ## Tax1bp1 Tax1bp1 ## Man2b1 Man2b1 ## Nfic Nfic ## Foxa2 Foxa2 ## Stard3nl Stard3nl ## Gm52543 Gm52543 ## Itga21 Itga2 ## Tubb2b1 Tubb2b ## Chst8 Chst8 ## Ntan1 Ntan1 ## Ccdc50 Ccdc50 ## Zfp30 Zfp30 ## Zbed3 Zbed3 ## Fat4 Fat4 ## Nucb1 Nucb1 ## Pde1c Pde1c ## Psen21 Psen2 ## Tmem38a Tmem38a ## Rpl21 Rpl21 ## Gp1bb Gp1bb ## Hmgb1 Hmgb1 ## Ggh Ggh ## Septin5 Septin5 ## Prdx21 Prdx2 ## Plaat3 Plaat3 ## Rab39b Rab39b ## Aplp11 Aplp1 ## Raph1 Raph1 ## Hmgn5 Hmgn5 ## Tmem2561 Tmem256 ## Ceacam18 Ceacam18 ## Gm5148 Gm5148 ## Steap2 Steap2 ## Chd6 Chd6 ## Tspan131 Tspan13 ## Rpl7a Rpl7a ## Hsd17b13 Hsd17b13 ## 0610040J01Rik 0610040J01Rik ## Slc35b3 Slc35b3 ## Pde8a1 Pde8a ## Dnajc15 Dnajc15 ## Cox8a1 Cox8a ## Fah Fah ## Arpc1a1 Arpc1a ## Mrpl17 Mrpl17 ## Gm32995 Gm32995 ## Samd11 Samd11 ## Ap1s3 Ap1s3 ## Immp1l Immp1l ## Pck2 Pck2 ## Galr2 Galr2 ## Gm30247 Gm30247 ## Ern1 Ern1 ## Kank2 Kank2 ## Epha31 Epha3 ## Sec61g1 Sec61g ## Ap1ar Ap1ar ## Pgm2l1 Pgm2l1 ## Eef1g Eef1g ## Iftap Iftap ## Cobll1 Cobll1 ## Fam204a Fam204a ## Sec11a1 Sec11a ## Psma5 Psma5 ## Nagk Nagk ## Erg28 Erg28 ## Tmem45a Tmem45a ## Mthfs Mthfs ## Arhgef37 Arhgef37 ## Mir100hg Mir100hg ## Nbea Nbea ## Gask1a Gask1a ## Manf Manf ## Vmac Vmac ## Tecr1 Tecr ## Caprin2 Caprin2 ## Pank1 Pank1 ## Ormdl21 Ormdl2 ## Rab27a Rab27a ## Pla2g15 Pla2g15 ## Serf1 Serf1 ## Pigp Pigp ## Sfr1 Sfr1 ## Frmd7 Frmd7 ## Selenoi Selenoi ## Rida Rida ## Creg11 Creg1 ## Adam32 Adam32 ## Rps9 Rps9 ## Rab11fip2 Rab11fip2 ## Gsto2 Gsto2 ## Nav2 Nav2 ## Rpl24 Rpl24 ## Mxd4 Mxd4 ## Gm2a Gm2a ## Per3 Per3 ## Calm31 Calm3 ## Lsr1 Lsr ## Hpgd Hpgd ## Lamb1 Lamb1 ## Selenow Selenow ## Ppdpf1 Ppdpf ## Trappc6b1 Trappc6b ## Sobp1 Sobp ## Lamtor21 Lamtor2 ## Sp3os1 Sp3os ## 2510009E07Rik 2510009E07Rik ## Ifit1 Ifit1 ## Antkmt1 Antkmt ## Slc35g21 Slc35g2 ## Rpl18a Rpl18a ## Agtpbp1 Agtpbp1 ## Glrx5 Glrx5 ## Prkrip11 Prkrip1 ## Guk11 Guk1 ## Med12 Med12 ## Stxbp11 Stxbp1 ## Ndufa71 Ndufa7 ## Btf31 Btf3 ## Ccdc169 Ccdc169 ## Ehd3 Ehd3 ## Cd99l2 Cd99l2 ## Mturn Mturn ## Abcc5 Abcc5 ## Krtcap2 Krtcap2 ## P3h4 P3h4 ## Smim27 Smim27 ## Tbca Tbca ## Chst12 Chst12 ## Cul1 Cul1 ## Rplp2 Rplp2 ## Spp2 Spp2 ## Gm34194 Gm34194 ## Cdhr2 Cdhr2 ## Krtcap3 Krtcap3 ## Ten1 Ten1 ## Isoc2b Isoc2b ## Ms4a5 Ms4a5 ## Eci2 Eci2 ## Rps21 Rps21 ## Gm41676 Gm41676 ## Asxl3 Asxl3 ## 2610035D17Rik1 2610035D17Rik ## Crybb1 Crybb1 ## Fndc1 Fndc1 ## Slc44a3 Slc44a3 ## Peg131 Peg13 ## Cnot8 Cnot8 ## Actn11 Actn1 ## Zfp697 Zfp697 ## Tmx4 Tmx4 ## Nfia Nfia ## Tmem107 Tmem107 ## Tmem30b1 Tmem30b ## Mapk8ip2 Mapk8ip2 ## Jade2 Jade2 ## Ceacam2 Ceacam2 ## Flot1 Flot1 ## Ndufa5 Ndufa5 ## Gipc1 Gipc1 ## Ssr2 Ssr2 ## Tpm41 Tpm4 ## Tdrkh Tdrkh ## Dynlt31 Dynlt3 ## Npdc11 Npdc1 ## Uqcr111 Uqcr11 ## Ncbp2as2 Ncbp2as2 ## Tm4sf51 Tm4sf5 ## Fech Fech ## Mif1 Mif ## Unc80 Unc80 ## Nipal1 Nipal1 ## Cxxc51 Cxxc5 ## Galns Galns ## Pon3 Pon3 ## Appl1 Appl1 ## Klf2 Klf2 ## Unc5c Unc5c ## Cops91 Cops9 ## Prdx6 Prdx6 ## Map1b Map1b ## Tbrg1 Tbrg1 ## Kif5b1 Kif5b ## Hexa1 Hexa ## Mamld1 Mamld1 ## Dnajc121 Dnajc12 ## Saraf Saraf ## Ppp1r14b Ppp1r14b ## Cbx6 Cbx6 ## Rabac11 Rabac1 ## Ndufs51 Ndufs5 ## Fgf1 Fgf1 ## Gm31471 Gm31471 ## Jpt2 Jpt2 ## Esd Esd ## Ppa1 Ppa1 ## Map2k6 Map2k6 ## Casp1 Casp1 ## Cebpzos Cebpzos ## Dpm31 Dpm3 ## Txndc15 Txndc15 ## Elavl4 Elavl4 ## Xkr7 Xkr7 ## Hes61 Hes6 ## Rnasek1 Rnasek ## Rps26 Rps26 ## Pdk31 Pdk3 ## C230057M02Rik C230057M02Rik ## Ybx1 Ybx1 ## Lamc2 Lamc2 ## Ntn4 Ntn4 ## C330008A17Rik C330008A17Rik ## Asap3 Asap3 ## Garin5a Garin5a ## Hsbp11 Hsbp1 ## Kif12 Kif12 ## Gm132031 Gm13203 ## Gc Gc ## Psip1 Psip1 ## Npc2 Npc2 ## Cpt2 Cpt2 ## Chchd4 Chchd4 ## Pgls Pgls ## Xist Xist ## Serpinb9 Serpinb9 ## Alg5 Alg5 ## Pgghg Pgghg ## Xkr6 Xkr6 ## Atraid Atraid ## Uqcc21 Uqcc2 ## Ttc39b1 Ttc39b ## Tmem191 Tmem191 ## Higd2a1 Higd2a ## Amigo1 Amigo1 ## Gfpt1 Gfpt1 ## Lingo1 Lingo1 ## 3110040N11Rik 3110040N11Rik ## Eif1ax Eif1ax ## Kcnh2 Kcnh2 ## Hdac5 Hdac5 ## Mrfap11 Mrfap1 ## Frk Frk ## Gm53610 Gm53610 ## Mrpl41 Mrpl41 ## Ggct Ggct ## Tusc31 Tusc3 ## 2900076A07Rik 2900076A07Rik ## Dhx32 Dhx32 ## Fam114a1 Fam114a1 ## Vdac3 Vdac3 ## Tiparp Tiparp ## Dph6 Dph6 ## Idnk Idnk ## Tmem71 Tmem71 ## Ypel2 Ypel2 ## Sec61b Sec61b ## Gm33502 Gm33502 ## Commd7 Commd7 ## Ppm1l Ppm1l ## Ooep Ooep ## Smim14 Smim14 ## Ddb11 Ddb1 ## Hmgn2 Hmgn2 ## Ifi47 Ifi47 ## Clic51 Clic5 ## Hap1 Hap1 ## Akr1e1 Akr1e1 ## Nedd81 Nedd8 ## Spmip3 Spmip3 ## Limd2 Limd2 ## Rps17 Rps17 ## Gfra2 Gfra2 ## Golm2 Golm2 ## Kctd10 Kctd10 ## Celf6 Celf6 ## Inpp1 Inpp1 ## Sytl5 Sytl5 ## Dnajc18 Dnajc18 ## Tmem266 Tmem266 ## Gm40109 Gm40109 ## Cep85 Cep85 ## Dram2 Dram2 ## Sec621 Sec62 ## Kras1 Kras ## Tmtc4 Tmtc4 ## Spcs1 Spcs1 ## Ndufc2 Ndufc2 ## Ntaq1 Ntaq1 ## Mtfp1 Mtfp1 ## Mapk15 Mapk15 ## Evl Evl ## Syp1 Syp ## Fdps1 Fdps ## Flywch21 Flywch2 ## Dap3 Dap3 ## Ndufa131 Ndufa13 ## Rps27l1 Rps27l ## Acbd6 Acbd6 ## Gcnt2 Gcnt2 ## Mast2 Mast2 ## Gm35835 Gm35835 ## Gja10 Gja10 ## Ndufb21 Ndufb2 ## Borcs71 Borcs7 ## Snhg8 Snhg8 ## Celf4 Celf4 ## Zfp6671 Zfp667 ## Gm41964 Gm41964 ## Dusp6 Dusp6 ## Klhdc8a1 Klhdc8a ## Neurl1a Neurl1a ## Ube2s Ube2s ## Eif3k1 Eif3k ## Selenof Selenof ## Sstr51 Sstr5 ## Zfp507 Zfp507 ## Tiprl Tiprl ## Fam234b Fam234b ## Pigs1 Pigs ## 2010004M13Rik 2010004M13Rik ## Mzt2 Mzt2 ## Copa1 Copa ## Zfp9381 Zfp938 ## Lage3 Lage3 ## Rb1 Rb1 ## Gm39467 Gm39467 ## Gm9908 Gm9908 ## Gstm6 Gstm6 ## Pla2g1b Pla2g1b ## Rpl5 Rpl5 ## Camk1 Camk1 ## Ina Ina ## Pitpnc1 Pitpnc1 ## Dctn31 Dctn3 ## Arpc31 Arpc3 ## Gmpr Gmpr ## Psenen1 Psenen ## Lppos Lppos ## Ugt2b35 Ugt2b35 ## Tmem591 Tmem59 ## Atp5f1b Atp5f1b ## Kxd1 Kxd1 ## Akr1c19 Akr1c19 ## Adgrg41 Adgrg4 ## Gm41804 Gm41804 ## Gm9926 Gm9926 ## Mthfsl1 Mthfsl ## Fam241b Fam241b ## Tns2 Tns2 ## Ifi301 Ifi30 ## Ruvbl1 Ruvbl1 ## Wdr83os Wdr83os ## Zfp945 Zfp945 ## Gng101 Gng10 ## Tnfsf12 Tnfsf12 ## Wfdc2 Wfdc2 ## Usp24 Usp24 ## Gm46528 Gm46528 ## Rundc3b Rundc3b ## Tecpr11 Tecpr1 ## Mag Mag ## Paip21 Paip2 ## Adh51 Adh5 ## Smim11 Smim11 ## Eid11 Eid1 ## Mrpl521 Mrpl52 ## Fam89a Fam89a ## Dctn6 Dctn6 ## Ubl51 Ubl5 ## Muc13 Muc13 ## Glra3 Glra3 ## Snhg4 Snhg4 ## Naca Naca ## Sytl2 Sytl2 ## Zfp709 Zfp709 ## Kcnh61 Kcnh6 ## Ift25 Ift25 ## Mapre2 Mapre2 ## Zfp961 Zfp961 ## Rpl23a Rpl23a ## Gusb1 Gusb ## Hnrnpa0 Hnrnpa0 ## Pdcd7 Pdcd7 ## Septin21 Septin2 ## Ptov11 Ptov1 ## Inpp5f Inpp5f ## Prorsd1 Prorsd1 ## Emc71 Emc7 ## Rhoa1 Rhoa ## Rraga1 Rraga ## Klhdc21 Klhdc2 ## Pik3c2b Pik3c2b ## Gm51641 Gm51641 ## Cntnap5b Cntnap5b ## Orm1 Orm1 ## Cyp4a32 Cyp4a32 ## Dynlrb11 Dynlrb1 ## Cryba21 Cryba2 ## Spg21 Spg21 ## Anp32e Anp32e ## Ccl27a Ccl27a ## Snrpf Snrpf ## Krt4 Krt4 ## Erp291 Erp29 ## Sugt11 Sugt1 ## Slc37a3 Slc37a3 ## Pofut2 Pofut2 ## C230034O21Rik C230034O21Rik ## Dcxr Dcxr ## Mrps18c1 Mrps18c ## Fahd1 Fahd1 ## Izumo4 Izumo4 ## Pin1 Pin1 ## Gm15706 Gm15706 ## Cd91 Cd9 ## Fahd2a Fahd2a ## A630066F11Rik A630066F11Rik ## Cirbp Cirbp ## Rpl26 Rpl26 ## Rpa3 Rpa3 ## Stmp1 Stmp1 ## Fcna Fcna ## Foxo1 Foxo1 ## Sdhaf4 Sdhaf4 ## Gm38487 Gm38487 ## Dusp23 Dusp23 ## Ppp1r9a Ppp1r9a ## Clic11 Clic1 ## Far1 Far1 ## Armcx4 Armcx4 ## Rnf111 Rnf11 ## Tprg1l1 Tprg1l ## Smim10l11 Smim10l1 ## Erlec1 Erlec1 ## Vsig1 Vsig1 ## Czib Czib ## Coq7 Coq7 ## Lamc1 Lamc1 ## Nefm Nefm ## Spock2 Spock2 ## Ccdc921 Ccdc92 ## Abhd14a Abhd14a ## Tigd2 Tigd2 ## Rtl8b Rtl8b ## Emb Emb ## Lipt2 Lipt2 ## Gstm5 Gstm5 ## Apool Apool ## Clic6 Clic6 ## Naxd1 Naxd ## Cimap1b Cimap1b ## Ucp21 Ucp2 ## Ufc11 Ufc1 ## Tbpl1 Tbpl1 ## Mtln Mtln ## Psmb8 Psmb8 ## Morf4l11 Morf4l1 ## Prnp1 Prnp ## Dnajc4 Dnajc4 ## Axdnd1 Axdnd1 ## As3mt As3mt ## Aopep Aopep ## Dnajc19 Dnajc19 ## Nek7 Nek7 ## Mrps16 Mrps16 ## Capn2 Capn2 ## Gm12146 Gm12146 ## Norad1 Norad ## Pet1001 Pet100 ## Spr Spr ## Ctbs Ctbs ## 6330407A03Rik 6330407A03Rik ## Tox1 Tox ## Lrrc58 Lrrc58 ## Tmem230 Tmem230 ## Psmg41 Psmg4 ## Ints6l Ints6l ## Zdhhc141 Zdhhc14 ## Snx6 Snx6 ## Gm53896 Gm53896 ## Sdf2l1 Sdf2l1 ## Rab9 Rab9 ## Dcaf8 Dcaf8 ## Trp53i111 Trp53i11 ## Tmem106c Tmem106c ## Psmb101 Psmb10 ## Gas21 Gas2 ## Aimp11 Aimp1 ## Ankrd13d Ankrd13d ## Cdkl2 Cdkl2 ## Slc25a36 Slc25a36 ## Tmem150b Tmem150b ## Rps27 Rps27 ## Arl6 Arl6 ## Map3k151 Map3k15 ## Med21 Med21 ## Xpnpep21 Xpnpep2 ## Colgalt2 Colgalt2 ## Gm7361 Gm7361 ## Psme11 Psme1 ## Ctss Ctss ## Jpx Jpx ## Nudcd2 Nudcd2 ## Porcn Porcn ## Rcn2 Rcn2 ## Hypk1 Hypk ## Gnai21 Gnai2 ## Dmd Dmd ## Lamtor41 Lamtor4 ## Trappc14 Trappc1 ## Ostf1 Ostf1 ## Rnf135 Rnf135 ## Cbfb Cbfb ## Cct5 Cct5 ## Cisd11 Cisd1 ## Cd1d1 Cd1d1 ## Bcas2 Bcas2 ## Meis2 Meis2 ## Ei241 Ei24 ## Tap2 Tap2 ## Sdhd Sdhd ## Homer1 Homer1 ## Lyn Lyn ## Tbcb1 Tbcb ## Rpp25l Rpp25l ## Med281 Med28 ## Arf4 Arf4 ## Lsm8 Lsm8 ## Pvt1 Pvt1 ## Ppp1r111 Ppp1r11 ## Ephx4 Ephx4 ## Sdcbp2 Sdcbp2 ## Smad11 Smad1 ## Sun3 Sun3 ## Vdac2 Vdac2 ## Rad51d Rad51d ## Fibp Fibp ## Lrp3 Lrp3 ## Psmb11 Psmb1 ## Atp8a11 Atp8a1 ## Zbp1 Zbp1 ## Mid2 Mid2 ## Micos13 Micos13 ## Tmed101 Tmed10 ## Eif11 Eif1 ## Scamp1 Scamp1 ## Cda Cda ## Fundc2 Fundc2 ## Eif2s21 Eif2s2 ## Arid5b Arid5b ## Zc3h12c Zc3h12c ## Hint2 Hint2 ## Prodh21 Prodh2 ## Psma21 Psma2 ## B9d1 B9d1 ## Gm26871 Gm26871 ## Mettl23 Mettl23 ## Atp5pf Atp5pf ## Eif3i Eif3i ## Ubb1 Ubb ## Bcl2 Bcl2 ## Rpp211 Rpp21 ## Bnip3l1 Bnip3l ## Angptl6 Angptl6 ## Ndufa8 Ndufa8 ## Foxa31 Foxa3 ## S100a131 S100a13 ## Glrx3 Glrx3 ## Cog11 Cog1 ## Gbp7 Gbp7 ## Dgcr6 Dgcr6 ## Gm15972 Gm15972 ## Csrp1 Csrp1 ## Jkamp1 Jkamp ## Churc11 Churc1 ## Ndufaf3 Ndufaf3 ## Vamp31 Vamp3 ## Hdac7 Hdac7 ## Ppargc1b Ppargc1b ## Nsf1 Nsf ## Cryl1 Cryl1 ## Gm10400 Gm10400 ## Katnal1 Katnal1 ## Tmem42 Tmem42 ## Sidt1 Sidt1 ## Mri1 Mri1 ## Ccdc126 Ccdc126 ## Cox6a11 Cox6a1 ## Nme31 Nme3 ## Ciao2b1 Ciao2b ## Gabbr2 Gabbr2 ## Akr1c131 Akr1c13 ## Tubb2a1 Tubb2a ## Lin37 Lin37 ## D130020L05Rik D130020L05Rik ## Hsd17b7 Hsd17b7 ## Gm28523 Gm28523 ## Gars1 Gars1 ## Gm32546 Gm32546 ## Pgap2 Pgap2 ## Atp8b5 Atp8b5 ## Tor3a1 Tor3a ## Naaa Naaa ## Lpcat2 Lpcat2 ## Mbnl21 Mbnl2 ## Gm39369 Gm39369 ## Rps6ka3 Rps6ka3 ## Rnf71 Rnf7 ## Zfp771 Zfp771 ## Ccdc30 Ccdc30 ## Ift1721 Ift172 ## Gimap5 Gimap5 ## Zfp422 Zfp422 ## Gm12576 Gm12576 ## Tmem1471 Tmem147 ## Nans Nans ## Oaz11 Oaz1 ## Snap471 Snap47 ## Samhd1 Samhd1 ## 4921528I07Rik 4921528I07Rik ## Mcrip11 Mcrip1 ## Emc9 Emc9 ## Cotl11 Cotl1 ## C230035I16Rik C230035I16Rik ## Cebpa Cebpa ## Pop7 Pop7 ## Trappc31 Trappc3 ## Ttll5 Ttll5 ## Marchf2 Marchf2 ## Swi5 Swi5 ## Glb11 Glb1 ## Ipo8 Ipo8 ## Fbxo211 Fbxo21 ## Nt5e1 Nt5e ## Eef1akmt1 Eef1akmt1 ## Armcx1 Armcx1 ## Cct31 Cct3 ## Tmbim4 Tmbim4 ## Dpy30 Dpy30 ## Pdcd51 Pdcd5 ## Mrpl531 Mrpl53 ## Cfdp1 Cfdp1 ## Parp14 Parp14 ## Tnks1bp1 Tnks1bp1 ## Svbp Svbp ## Tvp23b1 Tvp23b ## Atl3 Atl3 ## Il3ra Il3ra ## Fbxw17 Fbxw17 ## Ror2 Ror2 ## Gnaq Gnaq ## Rbpms Rbpms ## Ccdc86 Ccdc86 ## Ccpg1 Ccpg1 ## Rras1 Rras ## Pgbd5 Pgbd5 ## Mrpl571 Mrpl57 ## Fgf18 Fgf18 ## Scin Scin ## Smap2 Smap2 ## Pyurf Pyurf ## Macroh2a2 Macroh2a2 ## Casd1 Casd1 ## Csgalnact2 Csgalnact2 ## Ccdc28b Ccdc28b ## Nrp2 Nrp2 ## Rab25 Rab25 ## Mrpl43 Mrpl43 ## Fam216a Fam216a ## 1600014C10Rik 1600014C10Rik ## Gm41603 Gm41603 ## Sap18 Sap18 ## Ilk Ilk ## Msantd3 Msantd3 ## C1galt1c1 C1galt1c1 ## Fam221a Fam221a ## Pde2a1 Pde2a ## Rnf114 Rnf114 ## Eif5a Eif5a ## Tex30 Tex30 ## Tmem242 Tmem242 ## Lsm11 Lsm11 ## Unc79 Unc79 ## Cct71 Cct7 ## Polr2d Polr2d ## Mindy21 Mindy2 ## Rhob1 Rhob ## Slc12a5 Slc12a5 ## Wdfy2 Wdfy2 ## Ttc21b Ttc21b ## Ywhag1 Ywhag ## Cox6b11 Cox6b1 ## Acsf3 Acsf3 ## Ccdc107 Ccdc107 ## Cnot6l1 Cnot6l ## Chchd11 Chchd1 ## Sec61a11 Sec61a1 ## Ube2b Ube2b ## Itgb1bp1 Itgb1bp1 ## Polr2j1 Polr2j ## Fmc1 Fmc1 ## Gm16174 Gm16174 ## Lamtor5 Lamtor5 ## Nme11 Nme1 ## Eloc1 Eloc ## Rai2 Rai2 ## 4930594M22Rik 4930594M22Rik ## Ndufa41 Ndufa4 ## Hddc3 Hddc3 ## Ndufb5 Ndufb5 ## Ilkap1 Ilkap ## Ap2b11 Ap2b1 ## Tmie Tmie ## Uqcrh Uqcrh ## Gnaz1 Gnaz ## Bace11 Bace1 ## Arhgap6 Arhgap6 ## Trir1 Trir ## Cmc1 Cmc1 ## Borcs6 Borcs6 ## Gm34294 Gm34294 ## Phax Phax ## Uap1 Uap1 ## Rab3a1 Rab3a ## Xaf1 Xaf1 ## Trp53bp1 Trp53bp1 ## Paip2b Paip2b ## Gm10029 Gm10029 ## Cdc421 Cdc42 ## Psmg2 Psmg2 ## Ptrhd1 Ptrhd1 ## Baiap2l1 Baiap2l1 ## Phb1 Phb1 ## Arl1 Arl1 ## Tnr Tnr ## Dhrs7 Dhrs7 ## Abcc41 Abcc4 ## Mtmr71 Mtmr7 ## Dmxl2 Dmxl2 ## Cox5b Cox5b ## Selenom1 Selenom ## Glmn Glmn ## 1700066M21Rik 1700066M21Rik ## Adgrg7 Adgrg7 ## Tmem1651 Tmem165 ## Nop10 Nop10 ## Smim30 Smim30 ## Nap1l5 Nap1l5 ## Lpar5 Lpar5 ## Kbtbd12 Kbtbd12 ## Ddost Ddost ## D830035M03Rik D830035M03Rik ## 4933412E12Rik 4933412E12Rik ## Slc25a29 Slc25a29 ## Lamp21 Lamp2 ## Spata45 Spata45 ## 2310039H08Rik 2310039H08Rik ## Ctsz Ctsz ## Yy11 Yy1 ## Erp44 Erp44 ## Lhfpl4 Lhfpl4 ## Scmh1 Scmh1 ## Tmem151b Tmem151b ## Impdh11 Impdh1 ## Ramp2 Ramp2 ## Sh3yl11 Sh3yl1 ## Epsti1 Epsti1 ## Smim291 Smim29 ## Timm17a1 Timm17a ## Smpdl3a Smpdl3a ## Marchf91 Marchf9 ## Prrc1 Prrc1 ## Ube2h Ube2h ## Mocs1 Mocs1 ## Chchd6 Chchd6 ## Ndufb101 Ndufb10 ## Frmd6 Frmd6 ## Gm41753 Gm41753 ## Pcmt1 Pcmt1 ## Hras1 Hras ## Desi2 Desi2 ## Fes Fes ## F8a F8a ## Zfp81 Zfp81 ## Fndc3a Fndc3a ## Unc119 Unc119 ## Avpi1 Avpi1 ## Polr2g1 Polr2g ## Tle6 Tle6 ## Mvk Mvk ## Klhdc1 Klhdc1 ## Extl2 Extl2 ## Sort11 Sort1 ## Ing1 Ing1 ## Gnptg1 Gnptg ## Tmem2051 Tmem205 ## Gm45998 Gm45998 ## Hacd1 Hacd1 ## Tma71 Tma7 ## Arhgef9 Arhgef9 ## Laptm4a1 Laptm4a ## Anapc11 Anapc11 ## Arhgef10l Arhgef10l ## Nhlh2 Nhlh2 ## Mobp Mobp ## Cmya5 Cmya5 ## Vbp11 Vbp1 ## P3h1 P3h1 ## Dpyd Dpyd ## Mmp24 Mmp24 ## Atl2 Atl2 ## Cmbl Cmbl ## Zfp941 Zfp941 ## Adk Adk ## Bola3 Bola3 ## Gbp9 Gbp9 ## Ctps2 Ctps2 ## Tm7sf3 Tm7sf3 ## Zswim5 Zswim5 ## Igdcc4 Igdcc4 ## Fam229b Fam229b ## Prkaa11 Prkaa1 ## Sdhaf1 Sdhaf1 ## 1300002E11Rik 1300002E11Rik ## 2810001A02Rik 2810001A02Rik ## Cox7c Cox7c ## Setd31 Setd3 ## Ivd Ivd ## Tcp11 Tcp1 ## Ndrg4 Ndrg4 ## Ephb2 Ephb2 ## Gm14279 Gm14279 ## Dpy19l1 Dpy19l1 ## Lefty1 Lefty1 ## Dad11 Dad1 ## Dbndd2 Dbndd2 ## Hfe Hfe ## Zfp932 Zfp932 ## Sil1 Sil1 ## Mrpl201 Mrpl20 ## Hcar11 Hcar1 ## Usp8 Usp8 ## Slc66a3 Slc66a3 ## Drg11 Drg1 ## Rapgef4 Rapgef4 ## Zfp362 Zfp362 ## Prelid1 Prelid1 ## Gm54305 Gm54305 ## Gadd45g1 Gadd45g ## Serp21 Serp2 ## Acyp1 Acyp1 ## 2200002J24Rik 2200002J24Rik ## Rxylt1 Rxylt1 ## Bag4 Bag4 ## Ncstn1 Ncstn ## Mtfr1l Mtfr1l ## Gm16124 Gm16124 ## Tmem237 Tmem237 ## Dlat Dlat ## Mettl9 Mettl9 ## 2310009B15Rik 2310009B15Rik ## Rhou Rhou ## Mtif2 Mtif2 ## Hmg20a Hmg20a ## Btf3l4 Btf3l4 ## Hsd17b10 Hsd17b10 ## Edaradd Edaradd ## Ampd3 Ampd3 ## Gmfb1 Gmfb ## St18 St18 ## Armcx2 Armcx2 ## Ubxn1 Ubxn1 ## Kcna4 Kcna4 ## Ecd Ecd ## Gpr108 Gpr108 ## Socs1 Socs1 ## C1ra C1ra ## Ankra21 Ankra2 ## Fkbp3 Fkbp3 ## Aldh18a1 Aldh18a1 ## Gm38952 Gm38952 ## Klc3 Klc3 ## Sra11 Sra1 ## Thumpd1 Thumpd1 ## Adgrl2 Adgrl2 ## Gm19689 Gm19689 ## Slc23a2 Slc23a2 ## Tmem35b Tmem35b ## Srpk2 Srpk2 ## Txndc16 Txndc16 ## Tuba1a1 Tuba1a ## Gpr180 Gpr180 ## Ctxn1 Ctxn1 ## Pcmtd11 Pcmtd1 ## Rtraf Rtraf ## Gm54268 Gm54268 ## Zbtb7c Zbtb7c ## Fgl1 Fgl1 ## Cyp2c651 Cyp2c65 ## Gm51806 Gm51806 ## Timm10b1 Timm10b ## Atp5pb Atp5pb ## Gm44008 Gm44008 ## Msto1 Msto1 ## Mapk31 Mapk3 ## Coro2b Coro2b ## Haghl Haghl ## Prkar1a1 Prkar1a ## Prr15 Prr15 ## 1810008I18Rik 1810008I18Rik ## Kdelr21 Kdelr2 ## Sec22b Sec22b ## Pts Pts ## Trappc6a1 Trappc6a ## Hsdl2 Hsdl2 ## E2f2 E2f2 ## Emc101 Emc10 ## Gng2 Gng2 ## Timm231 Timm23 ## Spcs3 Spcs3 ## A1cf A1cf ## Mrps17 Mrps17 ## Crlf2 Crlf2 ## Ptpmt1 Ptpmt1 ## Med30 Med30 ## Abhd16a1 Abhd16a ## Erh Erh ## Tspyl4 Tspyl4 ## Lyrm9 Lyrm9 ## Rab11a1 Rab11a ## Prdx4 Prdx4 ## Nxn Nxn ## Zfp74 Zfp74 ## Tmem126a1 Tmem126a ## Def8 Def8 ## Ngrn Ngrn ## Tyw5 Tyw5 ## Rapgef5 Rapgef5 ## Gm12343 Gm12343 ## 1600027J07Rik 1600027J07Rik ## Atp6v0e21 Atp6v0e2 ## Nr1i2 Nr1i2 ## Bri3 Bri3 ## Smim19 Smim19 ## Slc9a7 Slc9a7 ## Bcor Bcor ## Cdkn1a Cdkn1a ## H2-T231 H2-T23 ## Gas7 Gas7 ## Ppme1 Ppme1 ## Qsox2 Qsox2 ## Mff Mff ## Alkbh6 Alkbh6 ## Gm33636 Gm33636 ## Gm343371 Gm34337 ## Bmyc Bmyc ## Lancl1 Lancl1 ## Triobp1 Triobp ## Ssna11 Ssna1 ## Gm465601 Gm46560 ## Scnm1 Scnm1 ## Tmem258 Tmem258 ## Nr2f2 Nr2f2 ## Ube2d2a1 Ube2d2a ## Tsnax Tsnax ## 2310022B05Rik 2310022B05Rik ## Pigyl Pigyl ## Lsm3 Lsm3 ## Frat2 Frat2 ## Cgn Cgn ## Zfp704 Zfp704 ## Myl12b1 Myl12b ## Zfp8711 Zfp871 ## Cd200 Cd200 ## Fem1b1 Fem1b ## Uqcc51 Uqcc5 ## Tm2d11 Tm2d1 ## Ntsr2 Ntsr2 ## 1500011B03Rik1 1500011B03Rik ## Cox14 Cox14 ## Eif4a2 Eif4a2 ## Lipc1 Lipc ## Slc44a11 Slc44a1 ## Rprd1a Rprd1a ## Myc Myc ## Fam83b Fam83b ## Tmed4 Tmed4 ## Atp2a21 Atp2a2 ## Cul7 Cul7 ## Pygb Pygb ## Ndufab1 Ndufab1 ## Gemin5 Gemin5 ## Tmem2381 Tmem238 ## Mtmr9 Mtmr9 ## Zdhhc17 Zdhhc17 ## Sigmar1 Sigmar1 ## Gtf2e2 Gtf2e2 ## Dbp Dbp ## Prmt11 Prmt1 ## Tmem14a Tmem14a ## Ldaf1 Ldaf1 ## Lekr1 Lekr1 ## Zfp318 Zfp318 ## Eid2 Eid2 ## Gal Gal ## Vamp21 Vamp2 ## Gm40743 Gm40743 ## 2310010J17Rik 2310010J17Rik ## Gtf2f1 Gtf2f1 ## Creb3 Creb3 ## Cmtm8 Cmtm8 ## Cyp3a16 Cyp3a16 ## Gm53937 Gm53937 ## Vps291 Vps29 ## Ino80d Ino80d ## Gal3st1 Gal3st1 ## Spryd7 Spryd7 ## Atp6ap11 Atp6ap1 ## Micu31 Micu3 ## Hint31 Hint3 ## Cep112 Cep112 ## Polb Polb ## Rbm43 Rbm43 ## Syt14 Syt14 ## Gnb21 Gnb2 ## Eef1akmt2 Eef1akmt2 ## Meiob Meiob ## Cnst Cnst ## Tmem276 Tmem276 ## Nudt10 Nudt10 ## Polr2c Polr2c ## Trappc2b Trappc2b ## Jtb Jtb ## Sar1a1 Sar1a ## Suclg1 Suclg1 ## A930038B10Rik1 A930038B10Rik ## Myl12a Myl12a ## Itfg11 Itfg1 ## Nbas1 Nbas ## Man2a2 Man2a2 ## Gpr85 Gpr85 ## Rex1bd1 Rex1bd ## Rin31 Rin3 ## Krt1 Krt1 ## Ninj1 Ninj1 ## Dll41 Dll4 ## Gm47095 Gm47095 ## Tenm2 Tenm2 ## R3hdm2 R3hdm2 ## Fdx1 Fdx1 ## Etv4 Etv4 ## Thap3 Thap3 ## Rufy2 Rufy2 ## Ldlrad4 Ldlrad4 ## Sumf2 Sumf2 ## Cadps1 Cadps ## Lyl1 Lyl1 ## Gm13594 Gm13594 ## Septin10 Septin10 ## Ppp2ca1 Ppp2ca ## Smim8 Smim8 ## Nkiras2 Nkiras2 ## Cby1 Cby1 ## Snrnp271 Snrnp27 ## Agpat5 Agpat5 ## Ebna1bp2 Ebna1bp2 ## Aph1b Aph1b ## H1f10 H1f10 ## Sgms1os1 Sgms1os1 ## Ndufb3 Ndufb3 ## Gabarapl1 Gabarapl1 ## Idi11 Idi1 ## Card191 Card19 ## Atp5mg1 Atp5mg ## Cd22 Cd22 ## Bola21 Bola2 ## Sar1b1 Sar1b ## Eef2kmt Eef2kmt ## Magoh Magoh ## Siah21 Siah2 ## Galnt31 Galnt3 ## Aldh6a1 Aldh6a1 ## Sacs Sacs ## Ss18l2 Ss18l2 ## Bloc1s2 Bloc1s2 ## Psph Psph ## Txndc121 Txndc12 ## Atp9a1 Atp9a ## Mycbp2 Mycbp2 ## Actr101 Actr10 ## Coq2 Coq2 ## Macrod2 Macrod2 ## Adcy2 Adcy2 ## Wdr6 Wdr6 ## Pcyox1l1 Pcyox1l ## Fam8a1 Fam8a1 ## Arl21 Arl2 ## Gm40922 Gm40922 ## Wdr83 Wdr83 ## 2410006H16Rik 2410006H16Rik ## Ttc331 Ttc33 ## H2az2 H2az2 ## Vgll4 Vgll4 ## Gm34147 Gm34147 ## Tyw1 Tyw1 ## Pcsk11 Pcsk1 ## Spg71 Spg7 ## Gm20208 Gm20208 ## F11r1 F11r ## Tmem161a Tmem161a ## Zmat21 Zmat2 ## 9030622O22Rik1 9030622O22Rik ## Cutal1 Cutal ## Zhx11 Zhx1 ## Ralgps1 Ralgps1 ## Auh Auh ## F21 F2 ## Akr1c121 Akr1c12 ## Timmdc1 Timmdc1 ## Utp3 Utp3 ## Mrps24 Mrps24 ## 2310015A10Rik 2310015A10Rik ## D730003I15Rik D730003I15Rik ## Synrg1 Synrg ## Cdk2ap11 Cdk2ap1 ## Il11ra1 Il11ra1 ## Foxo4 Foxo4 ## Psmb51 Psmb5 ## Unkl Unkl ## Pja21 Pja2 ## Luc7l2 Luc7l2 ## Gm41516 Gm41516 ## Dnajc21 Dnajc21 ## Gm30173 Gm30173 ## Gpd1l Gpd1l ## Ypel5 Ypel5 ## Smpd31 Smpd3 ## 1700110K17Rik 1700110K17Rik ## Gorasp21 Gorasp2 ## Plpp5 Plpp5 ## Ammecr1 Ammecr1 ## Dnajb141 Dnajb14 ## Tmem30a1 Tmem30a ## Rab13 Rab13 ## Fam3c1 Fam3c ## Hsbp1l1 Hsbp1l1 ## Chmp1b2 Chmp1b2 ## Galnt14 Galnt1 ## Gm51438 Gm51438 ## Vma211 Vma21 ## Eri3 Eri3 ## Gstm71 Gstm7 ## Cebpg1 Cebpg ## Dtna1 Dtna ## Dnase2a Dnase2a ## Oas1a Oas1a ## Usp291 Usp29 ## Adal Adal ## Neu2 Neu2 ## Glg11 Glg1 ## Pdia6 Pdia6 ## Btbd11 Btbd1 ## Cdc23 Cdc23 ## Rsu1 Rsu1 ## Cib2 Cib2 ## Pold2 Pold2 ## Slc66a21 Slc66a2 ## Ube2l6 Ube2l6 ## Pank31 Pank3 ## Mrpl12 Mrpl1 ## Fgf9 Fgf9 ## Edem2 Edem2 ## Pfdn2 Pfdn2 ## Hps1 Hps1 ## Dynlt2b Dynlt2b ## Sms1 Sms ## Gab1 Gab1 ## Nin Nin ## Adprh1 Adprh ## Bad Bad ## Zfp444 Zfp444 ## Sub11 Sub1 ## Rab3ip Rab3ip ## Sowahc1 Sowahc ## Mea11 Mea1 ## Surf1 Surf1 ## Atp5me1 Atp5me ## Fsd1l1 Fsd1l ## A030001D20Rik A030001D20Rik ## B230354K17Rik B230354K17Rik ## Cyp4v31 Cyp4v3 ## Med7 Med7 ## 2010320M18Rik 2010320M18Rik ## Asphd1 Asphd1 ## Samd9l Samd9l ## Gm46120 Gm46120 ## Igsf231 Igsf23 ## Tm2d21 Tm2d2 ## Nudt13 Nudt13 ## Trp53i13 Trp53i13 ## D330023K18Rik D330023K18Rik ## Psma31 Psma3 ## Tmem108 Tmem108 ## Gramd1a Gramd1a ## Limch1 Limch1 ## Got1 Got1 ## Arb2a1 Arb2a ## Gm39271 Gm39271 ## Fam3a Fam3a ## Rtl61 Rtl6 ## Echdc2 Echdc2 ## Eci1 Eci1 ## Smyd2 Smyd2 ## Smu1 Smu1 ## Xylt1 Xylt1 ## Vps251 Vps25 ## Ubac1 Ubac1 ## Cops8 Cops8 ## Tceal1 Tceal1 ## Psmb2 Psmb2 ## Orai3 Orai3 ## Dis3l2 Dis3l2 ## Cul5 Cul5 ## Pafah1b3 Pafah1b3 ## Rad21l Rad21l ## H4c4 H4c4 ## Smim26 Smim26 ## Sorbs2 Sorbs2 ## Enpp5 Enpp5 ## Znrd21 Znrd2 ## Atp6v1c11 Atp6v1c1 ## Bet1l Bet1l ## Rab211 Rab21 ## Mfsd10 Mfsd10 ## Kif3a Kif3a ## Efhd21 Efhd2 ## Alyref2 Alyref2 ## Zfp26 Zfp26 ## Pdia4 Pdia4 ## Pigbos1 Pigbos1 ## Slc17a5 Slc17a5 ## Arhgap11 Arhgap1 ## Ablim11 Ablim1 ## Mtg1 Mtg1 ## Bcl10 Bcl10 ## Rnh11 Rnh1 ## Nfe2l21 Nfe2l2 ## Miga1 Miga1 ## Degs1 Degs1 ## Ndufv2 Ndufv2 ## Fam120aos1 Fam120aos ## Trappc2 Trappc2 ## Serinc3 Serinc3 ## Cflar Cflar ## Faap20 Faap20 ## Os91 Os9 ## Commd61 Commd6 ## Qtrt1 Qtrt1 ## Letmd1 Letmd1 ## Rtl8a Rtl8a ## Calm11 Calm1 ## Gm44066 Gm44066 ## Atp1b3 Atp1b3 ## Nedd41 Nedd4 ## Spa17 Spa17 ## Scrn1 Scrn1 ## Rhobtb3 Rhobtb3 ## Vegfb Vegfb ## Amotl1 Amotl1 ## Lman1 Lman1 ## Zfpl1 Zfpl1 ## Ift27 Ift27 ## Dnaja41 Dnaja4 ## Lrba Lrba ## Plekhj1 Plekhj1 ## Tapbpl Tapbpl ## Irf1 Irf1 ## Sf3b5 Sf3b5 ## Ppp1ca1 Ppp1ca ## Carlr Carlr ## Halr1 Halr1 ## Atp8b3 Atp8b3 ## Gm42060 Gm42060 ## Gm32144 Gm32144 ## Igsf8 Igsf8 ## Ptpn9 Ptpn9 ## Smagp1 Smagp ## Slc25a11 Slc25a11 ## Slc39a10 Slc39a10 ## Atp8b2 Atp8b2 ## Sdf21 Sdf2 ## Tpd52l1 Tpd52l1 ## Iscu Iscu ## Pdia31 Pdia3 ## Zfp882 Zfp882 ## Gm34552 Gm34552 ## Fbxo3 Fbxo3 ## Rtn31 Rtn3 ## Lyrm2 Lyrm2 ## Cdkn2d Cdkn2d ## Hacd31 Hacd3 ## Tifa Tifa ## Psme2 Psme2 ## Zfand1 Zfand1 ## Slc43a21 Slc43a2 ## Srsf91 Srsf9 ## Mat2b Mat2b ## Rab6b1 Rab6b ## Mrpl22 Mrpl22 ## Slc39a61 Slc39a6 ## Snx21 Snx21 ## Irgm1 Irgm1 ## Rtn4rl11 Rtn4rl1 ## Lca5 Lca5 ## Med31 Med31 ## Patz1 Patz1 ## Taf101 Taf10 ## Gm15520 Gm15520 ## Gm6329 Gm6329 ## Nt5c3b Nt5c3b ## Chrac1 Chrac1 ## Rapgef3os2 Rapgef3os2 ## Tstd3 Tstd3 ## Chmp51 Chmp5 ## Spty2d1 Spty2d1 ## Stat5b Stat5b ## Apbb11 Apbb1 ## Mrps121 Mrps12 ## Arpc21 Arpc2 ## Maml3 Maml3 ## Gm33055 Gm33055 ## Vps36 Vps36 ## Tmem151a Tmem151a ## Arl15 Arl15 ## Pacs1 Pacs1 ## Ank3 Ank3 ## Herpud2 Herpud2 ## Txnl1 Txnl1 ## Surf4 Surf4 ## Anp32b1 Anp32b ## Kctd13 Kctd13 ## Cds2 Cds2 ## 2810001G20Rik 2810001G20Rik ## Dnmt3b Dnmt3b ## Furin Furin ## Naa38 Naa38 ## 2410022M11Rik 2410022M11Rik ## 4921524J17Rik 4921524J17Rik ## Mtch11 Mtch1 ## Tmem267 Tmem267 ## Iyd1 Iyd ## Hnrnph3 Hnrnph3 ## A930005H10Rik A930005H10Rik ## Slc25a23 Slc25a23 ## Ifit3 Ifit3 ## Gm54326 Gm54326 ## Ak1 Ak1 ## Etfb1 Etfb ## Vcf11 Vcf1 ## Sec22a1 Sec22a ## Eny2 Eny2 ## Cul4a Cul4a ## Triap1 Triap1 ## Txndc91 Txndc9 ## Snrpb Snrpb ## 1500026H17Rik 1500026H17Rik ## Tspan311 Tspan31 ## Uckl1 Uckl1 ## Ywhab1 Ywhab ## Slc30a7 Slc30a7 ## Rpl34-ps1 Rpl34-ps1 ## Srp91 Srp9 ## Dnajc1 Dnajc1 ## Rnf142 Rnf14 ## Rfx5 Rfx5 ## Gm10642 Gm10642 ## Hsd17b4 Hsd17b4 ## Tnks21 Tnks2 ## Cspg4b Cspg4b ## A930028N01Rik A930028N01Rik ## Kank3 Kank3 ## Mydgf1 Mydgf ## Amacr Amacr ## Tcta1 Tcta ## Syt17 Syt17 ## Pkm1 Pkm ## Slc39a13 Slc39a13 ## Eif1a Eif1a ## Gm52284 Gm52284 ## Lcmt1 Lcmt1 ## Miat Miat ## Grk21 Grk2 ## Gsdma1 Gsdma ## Naa20 Naa20 ## Abcc1 Abcc1 ## Ppp1r7 Ppp1r7 ## Btbd10 Btbd10 ## Chic2 Chic2 ## Tmem101 Tmem101 ## Oma1 Oma1 ## Tpp11 Tpp1 ## Copb21 Copb2 ## Zwint1 Zwint ## Gm33989 Gm33989 ## Angpt2 Angpt2 ## Rnaseh2c Rnaseh2c ## Sergef Sergef ## Ptma1 Ptma ## Adi11 Adi1 ## Naxe Naxe ## Nedd4l Nedd4l ## Irak1 Irak1 ## Map9 Map9 ## Taf13 Taf13 ## Grik5 Grik5 ## Pea15a1 Pea15a ## Snrnp35 Snrnp35 ## Phf10 Phf10 ## Dtymk Dtymk ## Drap11 Drap1 ## Ski Ski ## Jph4 Jph4 ## Xkr4 Xkr4 ## Nelfb Nelfb ## Zfand6 Zfand6 ## Nuak21 Nuak2 ## Psmd121 Psmd12 ## Rtcb Rtcb ## Rpp38 Rpp38 ## Snrpd3 Snrpd3 ## Hprt11 Hprt1 ## Vamp4 Vamp4 ## Echdc3 Echdc3 ## Atxn2 Atxn2 ## Yipf31 Yipf3 ## Gm38563 Gm38563 ## Cep19 Cep19 ## Eef2 Eef2 ## Gm4013 Gm4013 ## Tmem179b Tmem179b ## Usp14 Usp14 ## Rab42 Rab42 ## Phpt11 Phpt1 ## Fam149a1 Fam149a ## Nectin2 Nectin2 ## Klhl7 Klhl7 ## Trim27 Trim27 ## Ccdc136 Ccdc136 ## Cd83 Cd83 ## Zfp974 Zfp974 ## Cct8 Cct8 ## Ndufb61 Ndufb6 ## Ccdc88a Ccdc88a ## Fchsd2 Fchsd2 ## Eef1a2 Eef1a2 ## Anapc10 Anapc10 ## Mrpl34 Mrpl34 ## Spon2 Spon2 ## Cand11 Cand1 ## B4galt2 B4galt2 ## Gm52310 Gm52310 ## Map1lc3a1 Map1lc3a ## Pik3c2a Pik3c2a ## Ufm11 Ufm1 ## Diaph3 Diaph3 ## Enpp41 Enpp4 ## M6pr M6pr ## Cenpp Cenpp ## Ptger3 Ptger3 ## Nmt2 Nmt2 ## Loricrin Loricrin ## Trim3 Trim3 ## Lasp1 Lasp1 ## Trappc5 Trappc5 ## Psmd81 Psmd8 ## Bhlhb9 Bhlhb9 ## Gm26782 Gm26782 ## Gnb51 Gnb5 ## Ccs Ccs ## Rpl27 Rpl27 ## Snx14 Snx14 ## Zfp580 Zfp580 ## Eef1d Eef1d ## Wbp12 Wbp1 ## Pik3ip1 Pik3ip1 ## Tle2 Tle2 ## Gtf2a21 Gtf2a2 ## Pop51 Pop5 ## Leo1 Leo1 ## 2510002D24Rik 2510002D24Rik ## Epb41l4b1 Epb41l4b ## Mrpl28 Mrpl28 ## Polr2i1 Polr2i ## Dhx30 Dhx30 ## Gm38661 Gm38661 ## Yif1b Yif1b ## Camk2n21 Camk2n2 ## Fdft11 Fdft1 ## Lrrc31 Lrrc31 ## Wdr25 Wdr25 ## Prkca Prkca ## Adam101 Adam10 ## Itgb3bp Itgb3bp ## Rmdn3 Rmdn3 ## Zfp442 Zfp442 ## Ufsp2 Ufsp2 ## Kcnj16 Kcnj16 ## Sbsn Sbsn ## Vps281 Vps28 ## Gm16548 Gm16548 ## Dctn21 Dctn2 ## Adgrl1 Adgrl1 ## Mrpl301 Mrpl30 ## Vsig10 Vsig10 ## Lurap1l1 Lurap1l ## Taf11 Taf11 ## Kmt5a1 Kmt5a ## Ppa2 Ppa2 ## Zfp763 Zfp763 ## Gcsh Gcsh ## Atp6v0a11 Atp6v0a1 ## Daam11 Daam1 ## Msra Msra ## Rhoq Rhoq ## Nop16 Nop16 ## Gm4035 Gm4035 ## Taf9b Taf9b ## Mthfsd Mthfsd ## Nanp Nanp ## 1700108F19Rik 1700108F19Rik ## Alkbh7 Alkbh7 ## Kcnk11 Kcnk1 ## Tmem1641 Tmem164 ## Mrps341 Mrps34 ## Mrpl231 Mrpl23 ## Plxna3 Plxna3 ## Yeats41 Yeats4 ## Ramac1 Ramac ## C920006O11Rik C920006O11Rik ## Chil6 Chil6 ## Gm42094 Gm42094 ## Kcnb1 Kcnb1 ## Bbox1 Bbox1 ## Hook3 Hook3 ## Tnnt11 Tnnt1 ## Tnfaip8 Tnfaip8 ## Nae1 Nae1 ## Abca1 Abca1 ## Parp2 Parp2 ## Grpel2 Grpel2 ## Rabep11 Rabep1 ## Tsfm Tsfm ## Trpc1 Trpc1 ## Sel1l1 Sel1l ## Gale1 Gale ## 5430400D12Rik 5430400D12Rik ## Ogt Ogt ## Foxk1 Foxk1 ## Faah Faah ## Fyn1 Fyn ## 2700062C07Rik 2700062C07Rik ## Nicn1 Nicn1 ## Ap3s2 Ap3s2 ## Ces2b Ces2b ## Dtd2 Dtd2 ## Rrp11 Rrp1 ## Agbl3 Agbl3 ## Arpc51 Arpc5 ## Cerkl Cerkl ## Zfp707 Zfp707 ## Map3k3 Map3k3 ## Noc2l Noc2l ## Gata4 Gata4 ## Ppid Ppid ## Crybg3 Crybg3 ## Tmc41 Tmc4 ## Add1 Add1 ## Ankrd46 Ankrd46 ## Slc22a181 Slc22a18 ## Lsm4 Lsm4 ## Kit Kit ## Pomgnt11 Pomgnt1 ## Trmt1121 Trmt112 ## Pkig Pkig ## 1700007L15Rik 1700007L15Rik ## Lurap1 Lurap1 ## Cyld Cyld ## Ulk2 Ulk2 ## Cnrip1 Cnrip1 ## Dipk1a Dipk1a ## 5730455P16Rik 5730455P16Rik ## Rsph91 Rsph9 ## Gm34939 Gm34939 ## Abhd8 Abhd8 ## Wdfy4 Wdfy4 ## Gm15860 Gm15860 ## Gm13375 Gm13375 ## Cep83os Cep83os ## AI837181 AI837181 ## Sypl1 Sypl1 ## Lrp111 Lrp11 ## Naga1 Naga ## Lpp1 Lpp ## Cacna1h1 Cacna1h ## Hikeshi Hikeshi ## Ipp Ipp ## Fkbp4 Fkbp4 ## Gm34220 Gm34220 ## Timm17b Timm17b ## Pdia5 Pdia5 ## Zfp560 Zfp560 ## Ptgr21 Ptgr2 ## Arpc5l1 Arpc5l ## Gm51482 Gm51482 ## Atp5mc1 Atp5mc1 ## Tmtc1 Tmtc1 ## Cisd21 Cisd2 ## Ptprj Ptprj ## Rwdd2a Rwdd2a ## Akr1a11 Akr1a1 ## Tbc1d8b Tbc1d8b ## Cep15 Cep15 ## Ndufs41 Ndufs4 ## Yipf61 Yipf6 ## Snx5 Snx5 ## Adcy61 Adcy6 ## Mrpl40 Mrpl40 ## Ppp1cc1 Ppp1cc ## A230070E04Rik A230070E04Rik ## Trappc41 Trappc4 ## Hadhb Hadhb ## Pgam11 Pgam1 ## 1700054M17Rik 1700054M17Rik ## Gm37548 Gm37548 ## Sohlh2 Sohlh2 ## Gm833 Gm833 ## Gm46906 Gm46906 ## Gm46483 Gm46483 ## Tcp10c Tcp10c ## Ube2g2 Ube2g2 ## Lad1 Lad1 ## Cdk41 Cdk4 ## Ip6k21 Ip6k2 ## Hmox21 Hmox2 ## Hax11 Hax1 ## Smim71 Smim7 ## Zbtb14 Zbtb14 ## Saysd1 Saysd1 ## Tstd1 Tstd1 ## Tex2611 Tex261 ## Ube2d11 Ube2d1 ## Stoml2 Stoml2 ## Mitd11 Mitd1 ## Ccdc92b Ccdc92b ## Dpcd Dpcd ## Hadh Hadh ## Lrrcc1 Lrrcc1 ## Tgm5 Tgm5 ## Ftx Ftx ## 2310001H17Rik 2310001H17Rik ## Zfp82 Zfp82 ## Slc7a6 Slc7a6 ## Areg Areg ## Slc26a2 Slc26a2 ## Fam171a2 Fam171a2 ## Tmsb15b1 Tmsb15b1 ## Eif3m1 Eif3m ## Akirin2 Akirin2 ## Trmt10b Trmt10b ## Chchd21 Chchd2 ## Zdhhc12 Zdhhc12 ## Dcps Dcps ## Kifap3 Kifap3 ## Tdrd5 Tdrd5 ## 2310030G06Rik 2310030G06Rik ## Klhl36 Klhl36 ## Ncoa7 Ncoa7 ## Nipsnap11 Nipsnap1 ## Tmem132a Tmem132a ## Keap11 Keap1 ## Supt20 Supt20 ## Sdsl Sdsl ## Zfpm11 Zfpm1 ## Tsr11 Tsr1 ## Gm33682 Gm33682 ## Arhgap32 Arhgap32 ## Cers4 Cers4 ## Sac3d1 Sac3d1 ## Chmp2a1 Chmp2a ## 1110004F10Rik1 1110004F10Rik ## Mia2 Mia2 ## Pafah1b11 Pafah1b1 ## Plp2 Plp2 ## Med11 Med11 ## Dda11 Dda1 ## Diablo1 Diablo ## Sncg Sncg ## Poglut1 Poglut1 ## Sec11c Sec11c ## Muc20 Muc20 ## Prl6a1 Prl6a1 ## Armc1 Armc1 ## Gm46650 Gm46650 ## Cd46 Cd46 ## Tram1l1 Tram1l1 ## Gpr158 Gpr158 ## Mageh1 Mageh1 ## Gucd1 Gucd1 ## Ophn1 Ophn1 ## Slc39a7 Slc39a7 ## Ctf1 Ctf1 ## Dexi Dexi ## Zmpste241 Zmpste24 ## Slc35d1 Slc35d1 ## Tsix Tsix ## Pcgf2 Pcgf2 ## Klhdc10 Klhdc10 ## Dock9 Dock9 ## Tspan3 Tspan3 ## Stard101 Stard10 ## Cldnd1 Cldnd1 ## Ro601 Ro60 ## Pex3 Pex3 ## Cspg5 Cspg5 ## Eif3g1 Eif3g ## Vapa1 Vapa ## Cd274 Cd274 ## U2af1l41 U2af1l4 ## Gm32385 Gm32385 ## Ptpdc1 Ptpdc1 ## Zcrb1 Zcrb1 ## Gkap1 Gkap1 ## Gtf2i1 Gtf2i ## Arfgap1 Arfgap1 ## Onecut21 Onecut2 ## Selenoh Selenoh ## Bag5 Bag5 ## Sumo21 Sumo2 ## Sgtb Sgtb ## Mrps331 Mrps33 ## Pigg Pigg ## Tsen34 Tsen34 ## E130311K13Rik E130311K13Rik ## 2410002F23Rik 2410002F23Rik ## Dusp22 Dusp22 ## Polr3d Polr3d ## Rpl36al Rpl36al ## Gm10033 Gm10033 ## Smad2 Smad2 ## Gm35534 Gm35534 ## Gm2137 Gm2137 ## Fggy Fggy ## Sh3bgrl Sh3bgrl ## Rp9 Rp9 ## Pdhb Pdhb ## Elp3 Elp3 ## Zfp617 Zfp617 ## Isca2 Isca2 ## Gm320171 Gm32017 ## Dpm1 Dpm1 ## Mrpl13 Mrpl13 ## Tm9sf2 Tm9sf2 ## Zc3h7b1 Zc3h7b ## Gsk3b1 Gsk3b ## Gm36318 Gm36318 ## Gm40543 Gm40543 ## Atp5pd1 Atp5pd ## Nudt141 Nudt14 ## Kdm1b Kdm1b ## Cibar11 Cibar1 ## Lyset1 Lyset ## Ankrd24 Ankrd24 ## Psma11 Psma1 ## Brk11 Brk1 ## Ssr1 Ssr1 ## Mcf2l1 Mcf2l ## Xpa Xpa ## Actl6b Actl6b ## Med14 Med14 ## Ywhae1 Ywhae ## Tial1 Tial1 ## Thoc2 Thoc2 ## Nol12 Nol12 ## Pmvk Pmvk ## Rfx7 Rfx7 ## Ndufa111 Ndufa11 ## Asah11 Asah1 ## Prkx1 Prkx ## Rab141 Rab14 ## Ubap1l Ubap1l ## Gm45916 Gm45916 ## Chkb Chkb ## Slc17a9 Slc17a9 ## Fbxw9 Fbxw9 ## Abhd14b Abhd14b ## Zfp52 Zfp52 ## Polr2m1 Polr2m ## Gm40150 Gm40150 ## Slc39a11 Slc39a11 ## Fads11 Fads1 ## Tlcd4 Tlcd4 ## Rab321 Rab32 ## Fbxo17 Fbxo17 ## Chst11 Chst11 ## Gm32849 Gm32849 ## Cyren Cyren ## Bcl7a Bcl7a ## Gmds Gmds ## Ptprg1 Ptprg ## Rbx11 Rbx1 ## Tlcd3a1 Tlcd3a ## Fam83d1 Fam83d ## Selenon Selenon ## Nudt19 Nudt19 ## Dnajc31 Dnajc3 ## Ypel1 Ypel1 ## Tmem216 Tmem216 ## Gpr75 Gpr75 ## Cd79b Cd79b ## Dnajc30 Dnajc30 ## Gopc Gopc ## G630025P09Rik G630025P09Rik ## Chchd7 Chchd7 ## Rabggtb Rabggtb ## Pcdhb4 Pcdhb4 ## Itpripl2 Itpripl2 ## Gm46617 Gm46617 ## Cmc4 Cmc4 ## Palm Palm ## Zfp40 Zfp40 ## Acacb Acacb ## Golga71 Golga7 ## Slc25a12 Slc25a12 ## Mdc1 Mdc1 ## Pten1 Pten ## Gdap21 Gdap2 ## Abhd17a Abhd17a ## BC051226 BC051226 ## Krt101 Krt10 ## Hook1 Hook1 ## Fbxl20 Fbxl20 ## Cdh4 Cdh4 ## Mfsd4b1 Mfsd4b1 ## Naa35 Naa35 ## Smim20 Smim20 ## Abtb11 Abtb1 ## Tsc22d3 Tsc22d3 ## Id31 Id3 ## 0610040B10Rik 0610040B10Rik ## 4930503L19Rik 4930503L19Rik ## Wbp41 Wbp4 ## Gys1 Gys1 ## Pelp1 Pelp1 ## Herc3 Herc3 ## Gm15816 Gm15816 ## Zfp950 Zfp950 ## Nfu11 Nfu1 ## Ift74 Ift74 ## Fcor Fcor ## Suz12 Suz12 ## H2bc271 H2bc27 ## Msantd41 Msantd4 ## Suv39h1 Suv39h1 ## Dph3 Dph3 ## Anxa4 Anxa4 ## Kifbp Kifbp ## Ndufb71 Ndufb7 ## AI480526 AI480526 ## Tmem50a1 Tmem50a ## Cyp2j8 Cyp2j8 ## Dguok Dguok ## Gm36220 Gm36220 ## Ifih11 Ifih1 ## Polr2e1 Polr2e ## Hibch Hibch ## Spata24 Spata24 ## Gm35611 Gm35611 ## Ehd4 Ehd4 ## Nt5c1 Nt5c ## Arhgap29 Arhgap29 ## Cnn31 Cnn3 ## Znrf3 Znrf3 ## Ppp5c Ppp5c ## A930001C03Rik A930001C03Rik ## Lgals1 Lgals1 ## Lipa Lipa ## Aip1 Aip ## Pdk1 Pdk1 ## Gm7932 Gm7932 ## Acer1 Acer1 ## Pnkd1 Pnkd ## A430005L14Rik A430005L14Rik ## Uba21 Uba2 ## Thap111 Thap11 ## Manba Manba ## D730005E14Rik D730005E14Rik ## Cdk5rap3 Cdk5rap3 ## AW1120101 AW112010 ## Lsm5 Lsm5 ## Cfap52 Cfap52 ## Gatd1 Gatd1 ## C630043F03Rik C630043F03Rik ## Dnm1l Dnm1l ## Uggt2 Uggt2 ## Cnksr3 Cnksr3 ## Bcl11b Bcl11b ## Grb7 Grb7 ## Elk3 Elk3 ## Fkbp11 Fkbp11 ## Shroom1 Shroom1 ## Ttc32 Ttc32 ## Zfp593 Zfp593 ## Hcn3 Hcn3 ## 2210406O10Rik 2210406O10Rik ## Sft2d1 Sft2d1 ## Chid11 Chid1 ## Mrpl35 Mrpl35 ## Tmem250 Tmem250 ## Wwp1 Wwp1 ## Armcx3 Armcx3 ## Fgfbp3 Fgfbp3 ## Tnrc6b Tnrc6b ## Commd3 Commd3 ## Calhm21 Calhm2 ## Zdhhc8 Zdhhc8 ## Polr2f1 Polr2f ## Fam120c Fam120c ## Cops7a Cops7a ## Mfsd9 Mfsd9 ## Hilpda Hilpda ## Bccip Bccip ## Tmem141 Tmem141 ## Gm57587 Gm57587 ## Zfp637 Zfp637 ## Zfp551 Zfp551 ## Zmynd19 Zmynd19 ## Hipk3 Hipk3 ## Krcc11 Krcc1 ## A830021M18Rik A830021M18Rik ## Septin8 Septin8 ## Wrn Wrn ## Smarcb11 Smarcb1 ## Tsen15 Tsen15 ## Vps4b1 Vps4b ## Aldh7a11 Aldh7a1 ## Snhg10 Snhg10 ## Gm40656 Gm40656 ## Tm9sf41 Tm9sf4 ## Slc25a17 Slc25a17 ## Arhgef281 Arhgef28 ## Pdcd101 Pdcd10 ## Unc13a1 Unc13a ## Katnbl1 Katnbl1 ## Fkbp14 Fkbp14 ## Rwdd1 Rwdd1 ## Dcakd Dcakd ## Postn Postn ## Brdt Brdt ## Nrbp2 Nrbp2 ## Pdrg1 Pdrg1 ## Rps6ka2 Rps6ka2 ## Mppe1 Mppe1 ## Ifngr11 Ifngr1 ## Gatad2b Gatad2b ## Tmem135 Tmem135 ## Dnah7b Dnah7b ## Pds5b Pds5b ## Cep170b1 Cep170b ## Prr15l Prr15l ## Prpf6 Prpf6 ## Stat5a Stat5a ## Pam1 Pam ## Scand11 Scand1 ## Gm57735 Gm57735 ## Slc16a6 Slc16a6 ## 5430405H02Rik 5430405H02Rik ## Gemin71 Gemin7 ## Thoc7 Thoc7 ## Iah1 Iah1 ## Srgap2 Srgap2 ## Tacc1 Tacc1 ## Pde6d1 Pde6d ## Septin71 Septin7 ## Gm30789 Gm30789 ## Sema4d Sema4d ## Tsn1 Tsn ## Nudt11 Nudt11 ## Celf31 Celf3 ## Gm51920 Gm51920 ## Slc41a1 Slc41a1 ## Eea1 Eea1 ## Elp21 Elp2 ## 1700048O20Rik 1700048O20Rik ## Ift52 Ift52 ## Sod2 Sod2 ## Usp53 Usp53 ## Sh3glb11 Sh3glb1 ## Gm53687 Gm53687 ## Cep112it Cep112it ## Frs3 Frs3 ## Ggnbp2os Ggnbp2os ## Eml1 Eml1 ## Pink1 Pink1 ## 4930517O19Rik 4930517O19Rik ## Cap11 Cap1 ## Ciao2a1 Ciao2a ## Tada3 Tada3 ## Tmem60 Tmem60 ## Skic3 Skic3 ## Paupar Paupar ## Siah1a Siah1a ## Tmx3 Tmx3 ## Galnt11 Galnt11 ## Brix1 Brix1 ## Cops5 Cops5 ## Rab431 Rab43 ## Angel1 Angel1 ## Sdc1 Sdc1 ## Gm11963 Gm11963 ## Ankrd2 Ankrd2 ## Cox6c1 Cox6c ## Map3k12 Map3k12 ## Terf1 Terf1 ## 1810019D21Rik 1810019D21Rik ## Mkrn11 Mkrn1 ## Bola11 Bola1 ## Snx24 Snx24 ## Golga3 Golga3 ## Nup88 Nup88 ## Ripor1 Ripor1 ## Golt1a Golt1a ## Babam21 Babam2 ## Trafd1 Trafd1 ## Notch1 Notch1 ## Dancr Dancr ## Ddx50 Ddx50 ## Slain1 Slain1 ## Emc6 Emc6 ## Cct21 Cct2 ## 2610029K11Rik 2610029K11Rik ## Comt Comt ## Ube2j1 Ube2j1 ## Faf21 Faf2 ## Pafah1b21 Pafah1b2 ## Picalm Picalm ## Tmem260 Tmem260 ## Hnrnpr Hnrnpr ## Zfp3261 Zfp326 ## Rsl1d11 Rsl1d1 ## Gm49870 Gm49870 ## Zfp949 Zfp949 ## Cd59b1 Cd59b ## Fh1 Fh1 ## Ddx11 Ddx1 ## Gm53710 Gm53710 ## Tmem87b Tmem87b ## Sertad3 Sertad3 ## Pigf Pigf ## Gga2 Gga2 ## Kctd201 Kctd20 ## Mdp1 Mdp1 ## Ildr1 Ildr1 ## Tomm70a1 Tomm70a ## Nsfl1c1 Nsfl1c ## Adgb Adgb ## Dtx3 Dtx3 ## Cfap68 Cfap68 ## 4930579K19Rik 4930579K19Rik ## Usp48 Usp48 ## Mrps211 Mrps21 ## Gm17202 Gm17202 ## Cfl2 Cfl2 ## Enc1 Enc1 ## Mrps26 Mrps26 ## Gm17576 Gm17576 ## Gm36620 Gm36620 ## Usf1 Usf1 ## Sae1 Sae1 ## Wdr77 Wdr77 ## Mrpl14 Mrpl14 ## Rtn41 Rtn4 ## Add3 Add3 ## Mov10 Mov10 ## Vegfd Vegfd ## Plod3 Plod3 ## Adgrb3 Adgrb3 ## Oprl1 Oprl1 ## Gm46119 Gm46119 ## Crhbp Crhbp ## Gm53575 Gm53575 ## Gm53488 Gm53488 ## Usp13 Usp13 ## Gm16499 Gm16499 ## Ccdc179 Ccdc179 ## 9130015A21Rik 9130015A21Rik ## Gm34766 Gm34766 ## Adamts10 Adamts10 ## Cyp2c37 Cyp2c37 ## Tlr8 Tlr8 ## Slc25a47 Slc25a47 ## Uevld Uevld ## B9d2 B9d2 ## Stx1a1 Stx1a ## Extl3 Extl3 ## Bbln1 Bbln ## Psmb41 Psmb4 ## 2810408B13Rik 2810408B13Rik ## Spryd3 Spryd3 ## Ppp2r5a Ppp2r5a ## BC005624 BC005624 ## Enpp1 Enpp1 ## Bbip1 Bbip1 ## Gm39874 Gm39874 ## Arf5 Arf5 ## Trim34a Trim34a ## Trmt12 Trmt12 ## Gm19531 Gm19531 ## Mkrn2 Mkrn2 ## Camk2g Camk2g ## Osgep1 Osgep ## Acp21 Acp2 ## Actr8 Actr8 ## Gm35685 Gm35685 ## Rnf411 Rnf41 ## Pals2 Pals2 ## Phtf2 Phtf2 ## Cdkn2aipnl Cdkn2aipnl ## Uqcrb1 Uqcrb ## Ccdc1481 Ccdc148 ## Mid1 Mid1 ## Znhit3 Znhit3 ## Fam210b Fam210b ## Nipa1 Nipa1 ## Speer4c1 Speer4c1 ## Gorasp1 Gorasp1 ## Ergic11 Ergic1 ## Ube2q2 Ube2q2 ## E2f5 E2f5 ## Tmem2231 Tmem223 ## Kptn Kptn ## Rtn21 Rtn2 ## Phlda2 Phlda2 ## Smpd1 Smpd1 ## Ficd Ficd ## Capns11 Capns1 ## Prrx21 Prrx2 ## Inpp5e Inpp5e ## Dpp81 Dpp8 ## Slc35b21 Slc35b2 ## Zbtb33 Zbtb33 ## Slc16a12 Slc16a12 ## Anxa7 Anxa7 ## Prmt21 Prmt2 ## Ube2r21 Ube2r2 ## Ecscr Ecscr ## Tsr31 Tsr3 ## Olah Olah ## C1ql4 C1ql4 ## Gm4265 Gm4265 ## Insc Insc ## Hpcal4 Hpcal4 ## Gm31889 Gm31889 ## LOC118568708 LOC118568708 ## Nifk Nifk ## Pigk Pigk ## Ercc5 Ercc5 ## Tmem1601 Tmem160 ## Mark3 Mark3 ## Ice1 Ice1 ## Fank1 Fank1 ## Ssu721 Ssu72 ## Nit1 Nit1 ## Acap21 Acap2 ## Gm44148 Gm44148 ## Parl Parl ## Armh4 Armh4 ## Oxld1 Oxld1 ## Rad1 Rad1 ## Atp6v0b1 Atp6v0b ## Bbc3 Bbc3 ## Tle51 Tle5 ## Gm53915 Gm53915 ## Cnep1r11 Cnep1r1 ## Morf4l1b1 Morf4l1b ## Maip1 Maip1 ## Dok41 Dok4 ## 2510022D24Rik1 2510022D24Rik ## Dync1li2 Dync1li2 ## Gm57842 Gm57842 ## P4ha11 P4ha1 ## 5033403F01Rik 5033403F01Rik ## Cfap144 Cfap144 ## Mettl1 Mettl1 ## Bmf Bmf ## Ncbp21 Ncbp2 ## Susd6 Susd6 ## Exosc41 Exosc4 ## Gm26559 Gm26559 ## Bcl6 Bcl6 ## Gm36416 Gm36416 ## Dip2c Dip2c ## Ace31 Ace3 ## Ccdc591 Ccdc59 ## Atrnl11 Atrnl1 ## Gca Gca ## Gm53314 Gm53314 ## Mllt6 Mllt6 ## Gm11655 Gm11655 ## 1810044D09Rik 1810044D09Rik ## Ppp1r3c Ppp1r3c ## Mterf4 Mterf4 ## Cd200l11 Cd200l1 ## Atg5 Atg5 ## Mrpl50 Mrpl50 ## Evi5 Evi5 ## Tmem131l Tmem131l ## Coro7 Coro7 ## Nsa2 Nsa2 ## Sall4 Sall4 ## Rnf61 Rnf6 ## Vasp1 Vasp ## Usp37 Usp37 ## Rnf1411 Rnf141 ## Particl Particl ## Mllt111 Mllt11 ## Heatr5b Heatr5b ## Bet1 Bet1 ## Ints11 Ints11 ## Stx6 Stx6 ## Ube2n1 Ube2n ## Traf5 Traf5 ## Tmem86b1 Tmem86b ## Zc3h14 Zc3h14 ## Lrrc24 Lrrc24 ## Nab1 Nab1 ## 4632428C04Rik 4632428C04Rik ## Gm30084 Gm30084 ## Zfp821 Zfp821 ## Egln11 Egln1 ## Ccdc88c Ccdc88c ## Noa1 Noa1 ## Pycr3 Pycr3 ## 1110019D14Rik 1110019D14Rik ## Dcaf6 Dcaf6 ## Got2 Got2 ## Grn1 Grn ## Zfp955b Zfp955b ## Bbs5 Bbs5 ## Rtf2 Rtf2 ## Zcchc171 Zcchc17 ## R3hdm41 R3hdm4 ## Arfip11 Arfip1 ## Ints10 Ints10 ## Lgmn Lgmn ## 1110059G10Rik 1110059G10Rik ## Ssr31 Ssr3 ## Zc2hc1a Zc2hc1a ## Dtwd1 Dtwd1 ## Dgcr2 Dgcr2 ## Zbtb16 Zbtb16 ## Gm26588 Gm26588 ## Zbtb41 Zbtb41 ## Col8a1 Col8a1 ## Mfap31 Mfap3 ## Triqk1 Triqk ## Slc49a4 Slc49a4 ## Atp2c11 Atp2c1 ## Lman2l Lman2l ## Cbln31 Cbln3 ## Cog5 Cog5 ## Uba7 Uba7 ## Mindy11 Mindy1 ## Gm30871 Gm30871 ## Vamp81 Vamp8 ## Polr1h1 Polr1h ## Zkscan8 Zkscan8 ## Lama5 Lama5 ## Cpt1c Cpt1c ## Fuom Fuom ## Grsf11 Grsf1 ## Jmjd8 Jmjd8 ## 1110038F14Rik 1110038F14Rik ## Ufl1 Ufl1 ## Ttc1 Ttc1 ## Kdm7a1 Kdm7a ## Tmem231 Tmem231 ## Mnat1 Mnat1 ## Tmem167 Tmem167 ## Wbp21 Wbp2 ## Acot13 Acot13 ## Coprs1 Coprs ## Tor2a Tor2a ## Ipo4 Ipo4 ## Gpn3 Gpn3 ## Vkorc1l11 Vkorc1l1 ## Gm36180 Gm36180 ## Gm53213 Gm53213 ## Tmem175 Tmem175 ## Mapk1ip1 Mapk1ip1 ## Gm36026 Gm36026 ## Nln Nln ## Gm458711 Gm45871 ## Psmd31 Psmd3 ## Gm46915 Gm46915 ## Ambp1 Ambp ## Khdrbs3 Khdrbs3 ## Tcea2 Tcea2 ## Nectin4 Nectin4 ## Cdc371 Cdc37 ## Pycr1 Pycr1 ## Ift88 Ift88 ## Zfyve27 Zfyve27 ## 0610009E02Rik 0610009E02Rik ## Impa11 Impa1 ## Urod Urod ## Jade31 Jade3 ## Coa6 Coa6 ## Ptk71 Ptk7 ## Kcnd1 Kcnd1 ## Abhd17b Abhd17b ## Mid1ip11 Mid1ip1 ## Mesd Mesd ## Zfp51 Zfp51 ## Edc3 Edc3 ## A430046D13Rik A430046D13Rik ## Ptcra Ptcra ## Slc25a13 Slc25a1 ## Usp19 Usp19 ## Gnl3l1 Gnl3l ## Gm5113 Gm5113 ## Dusp7 Dusp7 ## Hspa51 Hspa5 ## Ttc22 Ttc22 ## Mterf2 Mterf2 ## Itga3 Itga3 ## Ubxn10 Ubxn10 ## 2310057M21Rik 2310057M21Rik ## Ube2w1 Ube2w ## Cnpy4 Cnpy4 ## Gm17753 Gm17753 ## Ccdc171 Ccdc171 ## Wdr47 Wdr47 ## Tasp1 Tasp1 ## B330016D10Rik B330016D10Rik ## Nova2 Nova2 ## Cryzl1 Cryzl1 ## Gm31356 Gm31356 ## Mgat4b Mgat4b ## Pfdn41 Pfdn4 ## Mrpl551 Mrpl55 ## Ric8a Ric8a ## H2aj1 H2aj ## Gar1 Gar1 ## Eif4ebp1 Eif4ebp1 ## Trim13 Trim13 ## G3bp21 G3bp2 ## Chst15 Chst15 ## Ccdc90b Ccdc90b ## Cyp20a11 Cyp20a1 ## Lpcat1 Lpcat1 ## Cdr2 Cdr2 ## Ykt61 Ykt6 ## Afap1 Afap1 ## Mpg Mpg ## Mettl25 Mettl25 ## Sdr39u1 Sdr39u1 ## Ndufb8 Ndufb8 ## Zfp523 Zfp523 ## Rnf21 Rnf2 ## Dr11 Dr1 ## Tdrd3 Tdrd3 ## Apoa2 Apoa2 ## Gm32164 Gm32164 ## Gm35574 Gm35574 ## Atxn3 Atxn3 ## Mrps28 Mrps28 ## Ppp3cc Ppp3cc ## Prkch Prkch ## Akr1b10 Akr1b10 ## Cuedc21 Cuedc2 ## Alad Alad ## Ddx421 Ddx42 ## Tmx21 Tmx2 ## Gm9902 Gm9902 ## Cers51 Cers5 ## Zfp788 Zfp788 ## Dnajb3 Dnajb3 ## Pip4k2b Pip4k2b ## Cstad Cstad ## Med10 Med10 ## Txlna Txlna ## Cbx8 Cbx8 ## Rhbdl1 Rhbdl1 ## Snhg12 Snhg12 ## Gtf3c61 Gtf3c6 ## Map4k4 Map4k4 ## Hpn Hpn ## Mrpl111 Mrpl11 ## Lsm7 Lsm7 ## Akap111 Akap11 ## Glod41 Glod4 ## Stt3a1 Stt3a ## Kantr Kantr ## Stox2 Stox2 ## Gjb11 Gjb1 ## Dph7 Dph7 ## Cbarp Cbarp ## Slc38a10 Slc38a10 ## Rit1 Rit1 ## Rubcn Rubcn ## Skic81 Skic8 ## Arf11 Arf1 ## Rab5b1 Rab5b ## Rbm101 Rbm10 ## Zfp280c Zfp280c ## Tor1a Tor1a ## Eif4a11 Eif4a1 ## Fam131c Fam131c ## Hopxos Hopxos ## Magohb Magohb ## Pla2g12b Pla2g12b ## Gramd4 Gramd4 ## Tent5a Tent5a ## Slc35a1 Slc35a1 ## Brip1os Brip1os ## Gpc4 Gpc4 ## Sec23b Sec23b ## Nutf21 Nutf2 ## Snrpb21 Snrpb2 ## Bckdk Bckdk ## Tusc2 Tusc2 ## Cep120 Cep120 ## Psmg1 Psmg1 ## Snx31 Snx3 ## Smurf1 Smurf1 ## Tgs1 Tgs1 ## Gm42250 Gm42250 ## Ube2l31 Ube2l3 ## Cyb5d1 Cyb5d1 ## Dhdds Dhdds ## Mob3a Mob3a ## Xpo7 Xpo7 ## Rgp1 Rgp1 ## Bub31 Bub3 ## Maneal Maneal ## B230219D22Rik1 B230219D22Rik ## Ppil21 Ppil2 ## Pou3f4 Pou3f4 ## Ankrd39 Ankrd39 ## Srd5a3 Srd5a3 ## Gm32031 Gm32031 ## Pnpla6 Pnpla6 ## Atpsckmt Atpsckmt ## Tmem109 Tmem109 ## Ddx31 Ddx31 ## Mettl4 Mettl4 ## Cyb5611 Cyb561 ## Exosc5 Exosc5 ## Mbd4 Mbd4 ## 1700040D17Rik 1700040D17Rik ## A930041C12Rik A930041C12Rik ## Fez1 Fez1 ## Uprt1 Uprt ## Pcif11 Pcif1 ## Capza21 Capza2 ## Ufd1 Ufd1 ## Scoc1 Scoc ## Gm29415 Gm29415 ## Tead41 Tead4 ## Hsd17b8 Hsd17b8 ## Pam161 Pam16 ## Pgm3 Pgm3 ## Gm54265 Gm54265 ## Ier3ip11 Ier3ip1 ## Gm39469 Gm39469 ## Ptch1 Ptch1 ## Lhpp Lhpp ## Pip4p1 Pip4p1 ## Anapc4 Anapc4 ## Rnf13 Rnf13 ## Gemin2 Gemin2 ## Chpf2 Chpf2 ## Gm38872 Gm38872 ## Ccnq Ccnq ## Arap1 Arap1 ## Jakmip11 Jakmip1 ## Npr11 Npr1 ## Dcaf71 Dcaf7 ## Cradd Cradd ## Srrm3 Srrm3 ## 4933434E20Rik 4933434E20Rik ## Yif1a1 Yif1a ## Cfap119 Cfap119 ## Dnal4 Dnal4 ## Fbxo8 Fbxo8 ## Rala1 Rala ## Tpcn1 Tpcn1 ## Gm53613 Gm53613 ## 4933421O10Rik 4933421O10Rik ## Phka2 Phka2 ## Ccm2 Ccm2 ## Srprb Srprb ## Sf3b31 Sf3b3 ## Cdk5 Cdk5 ## Glis2 Glis2 ## Gins4 Gins4 ## Gpam Gpam ## Dnajc17 Dnajc17 ## Card6 Card6 ## Malsu1 Malsu1 ## Mrto4 Mrto4 ## Tmed91 Tmed9 ## Zfp513 Zfp513 ## Nat8f1 Nat8f1 ## Psmb71 Psmb7 ## Msl11 Msl1 ## Yaf2 Yaf2 ## Eif3e Eif3e ## Senp8 Senp8 ## Gm348021 Gm34802 ## Ralb Ralb ## Zbtb42 Zbtb4 ## Gm32483 Gm32483 ## Twsg1 Twsg1 ## D630003M21Rik D630003M21Rik ## Foxk2 Foxk2 ## Pipox Pipox ## Dennd5a Dennd5a ## Zfp146 Zfp146 ## Ndfip11 Ndfip1 ## Dnaaf10 Dnaaf10 ## Gm156581 Gm15658 ## B4galt31 B4galt3 ## Rpap3 Rpap3 ## Abcc6 Abcc6 ## Nmb1 Nmb ## Acbd31 Acbd3 ## Rab19 Rab19 ## Entrep3 Entrep3 ## Cnp Cnp ## 1700102J08Rik 1700102J08Rik ## AI987944 AI987944 ## Fpgs Fpgs ## Uri1 Uri1 ## Gm26205 Gm26205 ## Fyttd11 Fyttd1 ## Rcor3 Rcor3 ## Gm32655 Gm32655 ## Denr1 Denr ## Mta3 Mta3 ## Dpm21 Dpm2 ## Cdk8 Cdk8 ## 4732491K20Rik 4732491K20Rik ## Gm49958 Gm49958 ## 5930430L01Rik 5930430L01Rik ## Gm57826 Gm57826 ## Bcap311 Bcap31 ## Sfrp5 Sfrp5 ## Mogs Mogs ## Mboat2 Mboat2 ## Cfap97 Cfap97 ## Tmem11 Tmem11 ## B3galnt2 B3galnt2 ## Tor4a Tor4a ## Mplkip Mplkip ## Fbxo32 Fbxo32 ## Kctd6 Kctd6 ## Cep164 Cep164 ## Wdr11 Wdr11 ## Yipf51 Yipf5 ## Clk3 Clk3 ## Ahi11 Ahi1 ## Zfp729b Zfp729b ## Zfp977 Zfp977 ## Rtbdn Rtbdn ## Rps6kl1 Rps6kl1 ## Cyp2c67 Cyp2c67 ## Oaz21 Oaz2 ## Borcs81 Borcs8 ## Capn7 Capn7 ## Zbtb8b Zbtb8b ## Rybp1 Rybp ## Gm12216 Gm12216 ## Mbtps1 Mbtps1 ## Stag2 Stag2 ## Ppm1n Ppm1n ## Mon1b Mon1b ## Srek1ip1 Srek1ip1 ## Dnajc221 Dnajc22 ## Ppt2 Ppt2 ## Cdv3 Cdv3 ## Mrm2 Mrm2 ## Cacng4 Cacng4 ## Mrpl32 Mrpl32 ## Il18bp Il18bp ## 4921531C22Rik 4921531C22Rik ## Fam219b Fam219b ## Josd21 Josd2 ## Dhx58 Dhx58 ## Eif2b3 Eif2b3 ## C2cd4c1 C2cd4c ## Tbc1d15 Tbc1d15 ## Psmd10 Psmd10 ## Timm9 Timm9 ## Tbck Tbck ## Dynlt1a Dynlt1a ## Elp4 Elp4 ## Sacm1l1 Sacm1l ## 9530082P21Rik 9530082P21Rik ## Ranbp91 Ranbp9 ## Zmat1 Zmat1 ## Ap1m21 Ap1m2 ## Cope Cope ## Clstn3 Clstn3 ## Lrrc201 Lrrc20 ## Ofd1 Ofd1 ## 2810029C07Rik 2810029C07Rik ## 2610306M01Rik1 2610306M01Rik ## Cdc16 Cdc16 ## Lrif1 Lrif1 ## Tapbp1 Tapbp ## Crem Crem ## Hipk1 Hipk1 ## Gng7 Gng7 ## Pnkp1 Pnkp ## Gm54251 Gm54251 ## Gm32633 Gm32633 ## Ntm Ntm ## Hebp2 Hebp2 ## Tbc1d23 Tbc1d23 ## Spag8 Spag8 ## 1110028F18Rik 1110028F18Rik ## Crb31 Crb3 ## Tbl1x1 Tbl1x ## Aldh5a1 Aldh5a1 ## Prima11 Prima1 ## Gm12996 Gm12996 ## Cstb1 Cstb ## Zfp1481 Zfp148 ## Kctd7 Kctd7 ## Cltb1 Cltb ## Lamtor31 Lamtor3 ## Glmp1 Glmp ## Cct6a1 Cct6a ## Slc39a3 Slc39a3 ## Gm14703 Gm14703 ## 9530062K07Rik 9530062K07Rik ## Actr31 Actr3 ## Gm30842 Gm30842 ## Acvr1 Acvr1 ## Arl8a1 Arl8a ## Adcy91 Adcy9 ## Klhl281 Klhl28 ## Zfp995 Zfp995 ## Sf3b61 Sf3b6 ## Sumo31 Sumo3 ## Trim21 Trim21 ## Cmc2 Cmc2 ## Scaper Scaper ## Leprot1 Leprot ## 1110006O24Rik 1110006O24Rik ## Gm39526 Gm39526 ## Anapc7 Anapc7 ## Plppr21 Plppr2 ## Prox1os Prox1os ## Zfp217 Zfp217 ## Jrk Jrk ## Snapin Snapin ## Oas2 Oas2 ## Ndufa61 Ndufa6 ## Ebag9 Ebag9 ## Rtkn Rtkn ## Trappc13 Trappc13 ## Apoo Apoo ## Caap1 Caap1 ## Sdk1 Sdk1 ## Arxes2 Arxes2 ## Pcyox11 Pcyox1 ## Sele Sele ## Gm5149 Gm5149 ## Cntfr Cntfr ## Frmpd1 Frmpd1 ## Gm35855 Gm35855 ## Dtx1 Dtx1 ## Gm53330 Gm53330 ## Gm36210 Gm36210 ## Nat1 Nat1 ## Gm27188 Gm27188 ## Gm10635 Gm10635 ## Rmst Rmst ## Kcnip1 Kcnip1 ## Rnf182 Rnf182 ## Gm34758 Gm34758 ## Gm41317 Gm41317 ## Gm53956 Gm53956 ## Gm53989 Gm53989 ## Gm47898 Gm47898 ## Entpd1 Entpd1 ## Gm32028 Gm32028 ## Il1rapl2 Il1rapl2 ## Gm15247 Gm15247 ## Rbm7 Rbm7 ## Clec11a Clec11a ## Cep41 Cep41 ## Sumo11 Sumo1 ## Fbxo7 Fbxo7 ## Zfp61 Zfp61 ## Tmem2191 Tmem219 ## Rab17 Rab17 ## Emc2 Emc2 ## Agap2 Agap2 ## Stx2 Stx2 ## Zfp72 Zfp72 ## Shroom2 Shroom2 ## Gm32440 Gm32440 ## Socs6 Socs6 ## Usp101 Usp10 ## Unc501 Unc50 ## Vcf2 Vcf2 ## Sh3bp41 Sh3bp4 ## Acot6 Acot6 ## Zfp791 Zfp791 ## Kdf1 Kdf1 ## Ubxn7 Ubxn7 ## Zfp189 Zfp189 ## 5031425F14Rik 5031425F14Rik ## Ermard Ermard ## Acad8 Acad8 ## Gpatch111 Gpatch11 ## Zkscan11 Zkscan1 ## Gm154111 Gm15411 ## Insig2 Insig2 ## Prkra Prkra ## Slain2 Slain2 ## Tgm7 Tgm7 ## Khdrbs2 Khdrbs2 ## Gm38985 Gm38985 ## Apoh Apoh ## Gm10447 Gm10447 ## Tspan33 Tspan33 ## Eprs1 Eprs1 ## 4930447K03Rik 4930447K03Rik ## Cyp1b1 Cyp1b1 ## D6Ertd527e D6Ertd527e ## Nwd1 Nwd1 ## Gm52101 Gm52101 ## 4833422M21Rik 4833422M21Rik ## Gm36587 Gm36587 ## Ocln1 Ocln ## Snhg17 Snhg17 ## Pard6a Pard6a ## Tmt1a Tmt1a ## Car11 Car11 ## Zfp13 Zfp13 ## AU022252 AU022252 ## Rec8 Rec8 ## Msh2 Msh2 ## Mllt3 Mllt3 ## Ptk2b Ptk2b ## Zfp429 Zfp429 ## Polr1g Polr1g ## Snu13 Snu13 ## Slc22a5 Slc22a5 ## Eif2ak2 Eif2ak2 ## Eif2s11 Eif2s1 ## Prr12 Prr12 ## Ap3m1 Ap3m1 ## Atp6v1e11 Atp6v1e1 ## Hpcal11 Hpcal1 ## Gm3740 Gm3740 ## Zmym3 Zmym3 ## Pdgfrl Pdgfrl ## Gm52313 Gm52313 ## Arsa Arsa ## Pierce2 Pierce2 ## Btbd6 Btbd6 ## Hdac3 Hdac3 ## Coasy Coasy ## Fbxo281 Fbxo28 ## Ext2 Ext2 ## Gm14399 Gm14399 ## Gm17224 Gm17224 ## Pigx Pigx ## Tox41 Tox4 ## Kpna3 Kpna3 ## Bod1 Bod1 ## Znrf1 Znrf1 ## Tmem50b1 Tmem50b ## Phf20 Phf20 ## Maea1 Maea ## Dusp31 Dusp3 ## Rgn Rgn ## Gpr52 Gpr52 ## Gspt2 Gspt2 ## Eid2b Eid2b ## Ahsa1 Ahsa1 ## Zfp414 Zfp414 ## Smim18 Smim18 ## Amer1 Amer1 ## Gm31775 Gm31775 ## Smim12 Smim12 ## Rinl Rinl ## Rps19bp11 Rps19bp1 ## Gm42027 Gm42027 ## Zfp605 Zfp605 ## Ift81 Ift81 ## Uqcc4 Uqcc4 ## Gm53355 Gm53355 ## Creld1 Creld1 ## Nalcn Nalcn ## Copb11 Copb1 ## Rfesd Rfesd ## Zxdb Zxdb ## Slc12a1 Slc12a1 ## Iqcb1 Iqcb1 ## Cyp4f13 Cyp4f13 ## D930003E18Rik D930003E18Rik ## 4930405A21Rik 4930405A21Rik ## Vnn3 Vnn3 ## Phactr3 Phactr3 ## Ldlr Ldlr ## D330022H12Rik D330022H12Rik ## Tmc3 Tmc3 ## Unc93b1 Unc93b1 ## Cstpp1 Cstpp1 ## 2210016L21Rik1 2210016L21Rik ## Mrpl44 Mrpl44 ## Gm31645 Gm31645 ## Elp51 Elp5 ## Fra10ac1 Fra10ac1 ## Pdap11 Pdap1 ## Zfp369 Zfp369 ## C230037L18Rik C230037L18Rik ## Nt5c2 Nt5c2 ## Synj2 Synj2 ## Degs21 Degs2 ## Zfp1008 Zfp1008 ## Zfp799 Zfp799 ## Xrcc51 Xrcc5 ## Ube2a1 Ube2a ## LTO1 LTO1 ## Tmed1 Tmed1 ## Haus21 Haus2 ## Srsf6 Srsf6 ## Gm30760 Gm30760 ## 4930526I15Rik 4930526I15Rik ## Gm39157 Gm39157 ## Cbr4 Cbr4 ## Igsf9b Igsf9b ## Rpl10 Rpl10 ## Clrn31 Clrn3 ## Mapk9 Mapk9 ## Bpnt21 Bpnt2 ## Prpf38a1 Prpf38a ## Sik2 Sik2 ## Smim131 Smim13 ## Tctn2 Tctn2 ## Nkapl Nkapl ## Qpctl Qpctl ## Ptprm Ptprm ## Terb1 Terb1 ## Gm36446 Gm36446 ## Cfap298 Cfap298 ## Maf1 Maf1 ## Chac2 Chac2 ## Nfkbil1 Nfkbil1 ## Tmem125 Tmem125 ## Mmp16 Mmp16 ## Gm30603 Gm30603 ## Snx12 Snx12 ## Gm37053 Gm37053 ## Surf2 Surf2 ## Trit1 Trit1 ## Gadd45gip11 Gadd45gip1 ## Cpped1 Cpped1 ## Pmpcb Pmpcb ## Gm54216 Gm54216 ## Myo1e1 Myo1e ## Uqcc6 Uqcc6 ## Ppp6c1 Ppp6c ## Gm34828 Gm34828 ## Tbc1d7 Tbc1d7 ## Gm46721 Gm46721 ## Rad9b Rad9b ## Lamtor11 Lamtor1 ## Ly96 Ly96 ## Nkiras1 Nkiras1 ## Cog7 Cog7 ## Mlkl Mlkl ## Bcdin3d Bcdin3d ## Pot1a Pot1a ## Smarcd11 Smarcd1 ## Txnrd2 Txnrd2 ## Vamp71 Vamp7 ## Blmh Blmh ## Sh3bgrl2 Sh3bgrl2 ## Gm38944 Gm38944 ## Pxn1 Pxn ## Rce11 Rce1 ## Magee1 Magee1 ## Wdr13 Wdr13 ## Klhl42 Klhl42 ## 2210416O15Rik 2210416O15Rik ## Gm1976 Gm1976 ## C130013N14Rik C130013N14Rik ## Cox15 Cox15 ## Polm Polm ## Tmem170 Tmem170 ## Sesn3 Sesn3 ## Hexd Hexd ## Gm13657 Gm13657 ## Rap2a Rap2a ## Zfp2601 Zfp260 ## Ddx60 Ddx60 ## Gm53283 Gm53283 ## Actr6 Actr6 ## Carnmt11 Carnmt1 ## Apmap1 Apmap ## Mta1 Mta1 ## Fbxo25 Fbxo25 ## Krba1 Krba1 ## Ift122 Ift122 ## Mmp23 Mmp23 ## Gm42149 Gm42149 ## Trappc11 Trappc11 ## Thumpd2 Thumpd2 ## Lrrc27 Lrrc27 ## Dnajb111 Dnajb11 ## Zmym1 Zmym1 ## Pik3c2g Pik3c2g ## Snhg3 Snhg3 ## Trim37 Trim37 ## Ogg1 Ogg1 ## Ndufs31 Ndufs3 ## Cdo1 Cdo1 ## Ppp1r15a Ppp1r15a ## P2ry61 P2ry6 ## Gabarap1 Gabarap ## Gm51556 Gm51556 ## Zfp810 Zfp810 ## Usp11 Usp11 ## 9130604C24Rik 9130604C24Rik ## Mknk2 Mknk2 ## Gm31793 Gm31793 ## Cnot9 Cnot9 ## Gdi21 Gdi2 ## Shc41 Shc4 ## Tymp Tymp ## Hace1 Hace1 ## Hspa2 Hspa2 ## Mknk1 Mknk1 ## Psmb61 Psmb6 ## Ppip5k21 Ppip5k2 ## Brms1 Brms1 ## Dclre1b Dclre1b ## Zscan21 Zscan21 ## Uhmk11 Uhmk1 ## Nif3l1 Nif3l1 ## Bmp6 Bmp6 ## Prss36 Prss36 ## Ddah1 Ddah1 ## Zfp775 Zfp775 ## Bmt2 Bmt2 ## Cbl Cbl ## Lmf1 Lmf1 ## Coq6 Coq6 ## Cdc731 Cdc73 ## Bax1 Bax ## Ankrd28 Ankrd28 ## Stau2 Stau2 ## Zfp119a Zfp119a ## Nphp3 Nphp3 ## Slc33a11 Slc33a1 ## Prrc2b1 Prrc2b ## Dph5 Dph5 ## Etaa1os Etaa1os ## Lrp8os2 Lrp8os2 ## Sin3b1 Sin3b ## Stip11 Stip1 ## Ric8b Ric8b ## Snhg6 Snhg6 ## Zfp661 Zfp661 ## Snd1 Snd1 ## 1110020A21Rik 1110020A21Rik ## Stambp Stambp ## Plekha4 Plekha4 ## Krt23 Krt23 ## Gm5431 Gm5431 ## Umps1 Umps ## Dlc1 Dlc1 ## Actr3b Actr3b ## Zbtb1 Zbtb1 ## Gm46410 Gm46410 ## Mrpl211 Mrpl21 ## Psma61 Psma6 ## Slc2a3 Slc2a3 ## Sspn Sspn ## Pfdn61 Pfdn6 ## Gm33447 Gm33447 ## Ly6f Ly6f ## Cmpk2 Cmpk2 ## Gm33130 Gm33130 ## Crls11 Crls1 ## Nck11 Nck1 ## Gm35601 Gm35601 ## Gm36790 Gm36790 ## Tmed21 Tmed2 ## Ap4b1 Ap4b1 ## Dtx2 Dtx2 ## Cebpz Cebpz ## 6030458C11Rik 6030458C11Rik ## Mrps23 Mrps23 ## Ercc1 Ercc1 ## Ythdf11 Ythdf1 ## Dhps Dhps ## Mtfr1 Mtfr1 ## Mthfd1 Mthfd1 ## Otulin Otulin ## G6pc3 G6pc3 ## Yae1d1 Yae1d1 ## Mrpl2 Mrpl2 ## Exosc1 Exosc1 ## Gm32998 Gm32998 ## Cox18 Cox18 ## Capn10 Capn10 ## Aida Aida ## Hhat Hhat ## Ndufaf7 Ndufaf7 ## Gm39807 Gm39807 ## Wdr89 Wdr89 ## Paqr7 Paqr7 ## Zhx3 Zhx3 ## Bst2 Bst2 ## B230217C12Rik B230217C12Rik ## Pim2 Pim2 ## Sinhcaf1 Sinhcaf ## Rnf122 Rnf122 ## 4930481A15Rik 4930481A15Rik ## Ring1 Ring1 ## Klhl12 Klhl12 ## Klf131 Klf13 ## Sipa1 Sipa1 ## B230369F24Rik B230369F24Rik ## Tor1aip11 Tor1aip1 ## Tmem170b Tmem170b ## Gm38521 Gm38521 ## Gm53861 Gm53861 ## Cd320 Cd320 ## Nsmce4a Nsmce4a ## Gm21145 Gm21145 ## Scn7a Scn7a ## Cyp2c38 Cyp2c38 ## Igfals Igfals ## Cnpy3 Cnpy3 ## Snx23 Snx2 ## Klhl91 Klhl9 ## Gm92221 Gm9222 ## Fam110c Fam110c ## Zfp280b Zfp280b ## Pgpep11 Pgpep1 ## Rnf1031 Rnf103 ## Fam32a1 Fam32a ## Rapgef1 Rapgef1 ## Adgre5 Adgre5 ## Nme4 Nme4 ## Gm13822 Gm13822 ## Cnnm3 Cnnm3 ## Cd86 Cd86 ## Hectd2os Hectd2os ## Samd101 Samd10 ## Fut4 Fut4 ## Tbl2 Tbl2 ## Cln5 Cln5 ## Gstp3 Gstp3 ## 5830448L01Rik 5830448L01Rik ## Pex11b Pex11b ## Slc9a61 Slc9a6 ## Mvd Mvd ## Ccdc181 Ccdc181 ## Zfp874a Zfp874a ## Gm14295 Gm14295 ## Btc Btc ## Fkbp81 Fkbp8 ## Eef1akmt4 Eef1akmt4 ## Zfp511 Zfp511 ## Golt1b Golt1b ## Rras21 Rras2 ## Tpd52l2 Tpd52l2 ## Npb Npb ## Mrnip Mrnip ## Poc1b1 Poc1b ## Dhrs13 Dhrs13 ## Fbxl161 Fbxl16 ## Manbal1 Manbal ## Kics2 Kics2 ## Ppp1r351 Ppp1r35 ## Wdr19 Wdr19 ## Gimap11 Gimap1 ## Gm43653 Gm43653 ## Toe1 Toe1 ## 2610528J11Rik1 2610528J11Rik ## Acot2 Acot2 ## Pgam5 Pgam5 ## Exoc8 Exoc8 ## Mpzl1 Mpzl1 ## Tgfa Tgfa ## Gm10190 Gm10190 ## Ydjc Ydjc ## Gm9776 Gm9776 ## Pdcd11 Pdcd11 ## Mcph1 Mcph1 ## Morn4 Morn4 ## Lmf2 Lmf2 ## Ttll12 Ttll12 ## Pspc1 Pspc1 ## C2cd4d C2cd4d ## Rnf139 Rnf139 ## Gm346861 Gm34686 ## Gpatch2l Gpatch2l ## Cep97 Cep97 ## Sike11 Sike1 ## Ccdc127 Ccdc127 ## Tgif1 Tgif1 ## Dnajc25 Dnajc25 ## Lancl2 Lancl2 ## Trim67 Trim67 ## Ncdn Ncdn ## Arrdc3 Arrdc3 ## Crbn1 Crbn ## Fbxw21 Fbxw2 ## Rad171 Rad17 ## Tmco11 Tmco1 ## Nap1l3 Nap1l3 ## Plin21 Plin2 ## Zfp780b Zfp780b ## F8 F8 ## Man2c11 Man2c1 ## Polr3g Polr3g ## Tgds Tgds ## Zbtb8os Zbtb8os ## Pskh1 Pskh1 ## Gm31728 Gm31728 ## Gm15489 Gm15489 ## Carm1 Carm1 ## Rab101 Rab10 ## Rpl7l11 Rpl7l1 ## Gm52134 Gm52134 ## Esam Esam ## Arpc1b1 Arpc1b ## Agt Agt ## Agps Agps ## 0610009L18Rik 0610009L18Rik ## Ttc5 Ttc5 ## E2f6 E2f6 ## Prelid2 Prelid2 ## Utp61 Utp6 ## 1810021B22Rik 1810021B22Rik ## Mypop Mypop ## Pdcd2l Pdcd2l ## Map1lc3b1 Map1lc3b ## Gm31619 Gm31619 ## Rab11bos1 Rab11bos1 ## Atn1 Atn1 ## Gm39499 Gm39499 ## Jmjd4 Jmjd4 ## Tmem70 Tmem70 ## 1110025M09Rik 1110025M09Rik ## Csad Csad ## 5830433I10Rik 5830433I10Rik ## Dqx1 Dqx1 ## Gm43568 Gm43568 ## Commd2 Commd2 ## Washc3 Washc3 ## Erfl Erfl ## Tomm40l Tomm40l ## Pabpn1l Pabpn1l ## Ube2d31 Ube2d3 ## Zfp60 Zfp60 ## Ndn Ndn ## 1700020N18Rik 1700020N18Rik ## Gm50595 Gm50595 ## Ccdc89 Ccdc89 ## Gm51715 Gm51715 ## Akr1c18 Akr1c18 ## Trim17 Trim17 ## Gm31305 Gm31305 ## Gm52781 Gm52781 ## Gm36223 Gm36223 ## Adamts9 Adamts9 ## Gm51458 Gm51458 ## F420014N23Rik F420014N23Rik ## Selenoo Selenoo ## Xpo4 Xpo4 ## Atf6b1 Atf6b ## Rasa11 Rasa1 ## Zfp78 Zfp78 ## Slc25a38 Slc25a38 ## Kcnh7 Kcnh7 ## Tsen2 Tsen2 ## Zbtb32 Zbtb32 ## Gm420671 Gm42067 ## Pcdhb5 Pcdhb5 ## Rmnd5b Rmnd5b ## Nsmf Nsmf ## Ebi3 Ebi3 ## Zc3h6 Zc3h6 ## Osbpl6 Osbpl6 ## Asnsd1 Asnsd1 ## Tbc1d19 Tbc1d19 ## Gm42253 Gm42253 ## Cep78 Cep78 ## Kdm4a Kdm4a ## Thnsl1 Thnsl1 ## Cldn61 Cldn6 ## Appbp21 Appbp2 ## Eola1 Eola1 ## Gm41231 Gm41231 ## 5830454E08Rik 5830454E08Rik ## Uck2 Uck2 ## Ttl Ttl ## B3gnt8 B3gnt8 ## Cfap300 Cfap300 ## Rpgrip1l Rpgrip1l ## Tefm Tefm ## Gm32013 Gm32013 ## Polr3gl Polr3gl ## Usp32 Usp32 ## Pno1 Pno1 ## Zfp111 Zfp111 ## Sfxn3 Sfxn3 ## Pcsk2os1 Pcsk2os1 ## AA388235 AA388235 ## Gna12 Gna12 ## Stx81 Stx8 ## Ubl4a Ubl4a ## Gm30906 Gm30906 ## Trim321 Trim32 ## Bscl2 Bscl2 ## Tmem17 Tmem17 ## Ccdc159 Ccdc159 ## Samd5 Samd5 ## Abhd10 Abhd10 ## Ksr2 Ksr2 ## Rbbp7 Rbbp7 ## Nup62 Nup62 ## Usp31 Usp31 ## Pold3 Pold3 ## Vsig10l Vsig10l ## Nup133 Nup133 ## Nosip Nosip ## 1700123O20Rik 1700123O20Rik ## Nudt1 Nudt1 ## Gm39342 Gm39342 ## Slc31a21 Slc31a2 ## Chchd5 Chchd5 ## Zfp2661 Zfp266 ## Pigu1 Pigu ## Eml2 Eml2 ## Trim141 Trim14 ## 9430065F17Rik 9430065F17Rik ## Rpusd4 Rpusd4 ## Rasl2-9 Rasl2-9 ## Becn11 Becn1 ## Fam20a1 Fam20a ## Tfpt Tfpt ## Gdf15 Gdf15 ## B3galt6 B3galt6 ## Rbm48 Rbm48 ## Pctp Pctp ## H131 H13 ## Tomm61 Tomm6 ## 9130401M01Rik 9130401M01Rik ## 4930556M19Rik 4930556M19Rik ## Gm57630 Gm57630 ## Klhl15 Klhl15 ## St6galnac4 St6galnac4 ## Gm42012 Gm42012 ## Agk Agk ## Pigc Pigc ## Mob3b Mob3b ## Prkaca1 Prkaca ## Gm7457 Gm7457 ## Gm4419 Gm4419 ## Haus7 Haus7 ## Zfp330 Zfp330 ## Pcbp4 Pcbp4 ## Gm3807 Gm3807 ## Zfp119b Zfp119b ## Champ1 Champ1 ## Ldlrap1 Ldlrap1 ## Gm26995 Gm26995 ## Zfp9 Zfp9 ## LOC102631992 LOC102631992 ## Gm53773 Gm53773 ## Tmem169 Tmem169 ## Stub11 Stub1 ## Gm51704 Gm51704 ## Gm26844 Gm26844 ## Gm52678 Gm52678 ## Lrrc73 Lrrc73 ## Adck2 Adck2 ## Gm46488 Gm46488 ## Atp6v0e1 Atp6v0e ## Snupn Snupn ## Ift140 Ift140 ## Prmt5 Prmt5 ## Galt Galt ## Gm46195 Gm46195 ## Gm39000 Gm39000 ## C920021L13Rik C920021L13Rik ## Bckdha Bckdha ## Pogz1 Pogz ## Gm14326 Gm14326 ## Nkap Nkap ## Memo1 Memo1 ## Fam185a Fam185a ## Mus81 Mus81 ## Gm46786 Gm46786 ## Ube2i1 Ube2i ## Get1 Get1 ## Tnfrsf14 Tnfrsf14 ## Bud23 Bud23 ## Clcn31 Clcn3 ## Vps8 Vps8 ## Cep131 Cep131 ## Zfp777 Zfp777 ## Gm52861 Gm52861 ## Zkscan7 Zkscan7 ## Dand5 Dand5 ## Cd99 Cd99 ## Stimate Stimate ## Trpv6 Trpv6 ## AW046200 AW046200 ## Gm43380 Gm43380 ## Nudt12 Nudt12 ## Zfp41 Zfp41 ## Thyn1 Thyn1 ## Ngef Ngef ## Gm53595 Gm53595 ## Rell2 Rell2 ## Gm10614 Gm10614 ## Thap1 Thap1 ## Prps1l31 Prps1l3 ## Ptgds Ptgds ## 1700026J14Rik 1700026J14Rik ## 4930579G18Rik 4930579G18Rik ## Kcnrg Kcnrg ## Tmem178 Tmem178 ## Gm33335 Gm33335 ## Akap14 Akap14 ## Alg2 Alg2 ## Gm25663 Gm25663 ## Gyg1 Gyg1 ## Cyp2d22 Cyp2d22 ## Pcdhga5 Pcdhga5 ## Oaz3 Oaz3 ## Cxcl9 Cxcl9 ## Gm40578 Gm40578 ## Gm5215 Gm5215 ## Trap1 Trap1 ## Gm34507 Gm34507 ## Rbm8a1 Rbm8a ## Gm33610 Gm33610 ## Gm52486 Gm52486 ## Gm53349 Gm53349 ## Gm53386 Gm53386 ## 4933427E11Rik 4933427E11Rik ## Ltbp3 Ltbp3 ## Aacs1 Aacs ## Brf2 Brf2 ## Zfp28 Zfp28 ## Fam53a Fam53a ## D7Ertd128e D7Ertd128e ## Vps26c1 Vps26c ## Cdin1 Cdin1 ## Fbxw81 Fbxw8 ## Psmd21 Psmd2 ## Bcap29 Bcap29 ## Ndufv31 Ndufv3 ## Gaa1 Gaa ## Qprt Qprt ## Map10 Map10 ## Ipo9 Ipo9 ## Gas8 Gas8 ## Pigt1 Pigt ## Dusp28 Dusp28 ## Zfp865 Zfp865 ## Zfp658 Zfp658 ## Gm6658 Gm6658 ## Tbcc1 Tbcc ## Rita1 Rita1 ## Ppox Ppox ## Zfp689 Zfp689 ## Gm356071 Gm35607 ## Fip1l1 Fip1l1 ## Dnajc9 Dnajc9 ## Mgat1 Mgat1 ## Gmpr2 Gmpr2 ## Mfhas1 Mfhas1 ## Gm33409 Gm33409 ## Ap4s1 Ap4s1 ## Tmem218 Tmem218 ## Gm14051 Gm14051 ## Rnaseh2a Rnaseh2a ## Hoxb2 Hoxb2 ## Cdc42ep41 Cdc42ep4 ## Prkd2 Prkd2 ## Pex26 Pex26 ## Ttll1 Ttll1 ## Hdac21 Hdac2 ## Coa4 Coa4 ## Dmap11 Dmap1 ## Tpgs2 Tpgs2 ## Gmppa Gmppa ## Phykpl Phykpl ## Kpna61 Kpna6 ## Selenbp1 Selenbp1 ## Thumpd3 Thumpd3 ## Ldhd Ldhd ## Nfatc2 Nfatc2 ## Mir703 Mir703 ## Gtpbp8 Gtpbp8 ## 2210417A02Rik1 2210417A02Rik ## Mnd1 Mnd1 ## Slc19a2 Slc19a2 ## Rela Rela ## Pemt Pemt ## Pcsk2 Pcsk2 ## Pum3 Pum3 ## Acp11 Acp1 ## Gm40448 Gm40448 ## Gm46941 Gm46941 ## Zkscan17 Zkscan17 ## 9330020H09Rik 9330020H09Rik ## 2010001A14Rik 2010001A14Rik ## Gm41408 Gm41408 ## Gosr1 Gosr1 ## Zfp1101 Zfp110 ## Nsdhl Nsdhl ## Zfp39 Zfp39 ## Reep2 Reep2 ## Kdm2b Kdm2b ## Ribc1 Ribc1 ## Parp3 Parp3 ## Gas2l3 Gas2l3 ## Gm46775 Gm46775 ## Ak6 Ak6 ## 6430503K07Rik 6430503K07Rik ## Purg Purg ## Bloc1s3 Bloc1s3 ## Gm10087 Gm10087 ## Zbtb34 Zbtb34 ## Lrfn1 Lrfn1 ## Osbpl8 Osbpl8 ## Pdp2 Pdp2 ## Dnajc16 Dnajc16 ## Rbm31 Rbm3 ## Ogdhl Ogdhl ## Tmem184c Tmem184c ## Pcnx4 Pcnx4 ## Fnbp1 Fnbp1 ## Tmem91 Tmem91 ## Katnb1 Katnb1 ## Map6 Map6 ## Gm8290 Gm8290 ## Carmil2 Carmil2 ## Epb42 Epb42 ## Gm41845 Gm41845 ## Tctn1 Tctn1 ## Gm4532 Gm4532 ## Aars2 Aars2 ## Mast1 Mast1 ## Prmt7 Prmt7 ## Clybl Clybl ## Cd247 Cd247 ## Zfp647 Zfp647 ## Rsph4a Rsph4a ## Ppp2r5b1 Ppp2r5b ## 5730409E04Rik 5730409E04Rik ## Eipr1 Eipr1 ## Cpsf3 Cpsf3 ## Fgd1 Fgd1 ## Fam199x1 Fam199x ## Svop Svop ## Pcdhga6 Pcdhga6 ## Pbx2 Pbx2 ## Gm5296 Gm5296 ## Prcp Prcp ## AB010352 AB010352 ## Septin9 Septin9 ## H2ap H2ap ## Tmem68 Tmem68 ## Ubfd1 Ubfd1 ## Ppm1g1 Ppm1g ## R3hcc1 R3hcc1 ## 2010315B03Rik 2010315B03Rik ## Gm16845 Gm16845 ## Dcaf12l1 Dcaf12l1 ## Kif3c Kif3c ## Fundc1 Fundc1 ## Tmem185b1 Tmem185b ## Megf8 Megf8 ## Snhg91 Snhg9 ## Gm5914 Gm5914 ## Klhl5 Klhl5 ## Brsk1 Brsk1 ## Zfp582 Zfp582 ## Bbs7 Bbs7 ## Catsper2 Catsper2 ## Invs Invs ## Parp11 Parp11 ## Crlf3 Crlf3 ## Meaf61 Meaf6 ## 1600012H06Rik 1600012H06Rik ## Rnaseh1 Rnaseh1 ## Isy1 Isy1 ## Gm40190 Gm40190 ## Bicdl2 Bicdl2 ## Cdyl Cdyl ## Gm30706 Gm30706 ## Mex3c Mex3c ## Mrps111 Mrps11 ## Atp5f1e1 Atp5f1e ## Zfp583 Zfp583 ## Tlnrd11 Tlnrd1 ## Zscan25 Zscan25 ## Dipk1b Dipk1b ## Mfsd14b1 Mfsd14b ## Gm52692 Gm52692 ## Il17ra Il17ra ## Hps6 Hps6 ## Gm22063 Gm22063 ## Chn11 Chn1 ## Plp1 Plp1 ## Zfp105 Zfp105 ## Or5m3b Or5m3b ## Gm32449 Gm32449 ## Dmkn Dmkn ## Ppih Ppih ## Casp9 Casp9 ## Slc2a8 Slc2a8 ## Pstk Pstk ## Gm22299 Gm22299 ## Tjap1 Tjap1 ## Bloc1s5 Bloc1s5 ## Gm15927 Gm15927 ## Gm33315 Gm33315 ## Herc6 Herc6 ## Mrpl39 Mrpl39 ## Kcnq4 Kcnq4 ## Zfp606 Zfp606 ## Eif2ak4 Eif2ak4 ## Avp Avp ## Gm38529 Gm38529 ## Adamtsl4 Adamtsl4 ## Lrrc8c Lrrc8c ## Kmo Kmo ## Gm33782 Gm33782 ## Eno1b Eno1b ## Gm13032 Gm13032 ## Cd72 Cd72 ## Prkab1 Prkab1 ## Acbd7 Acbd7 ## E130112N10Rik E130112N10Rik ## Gm53491 Gm53491 ## Zgpat Zgpat ## 4930448E22Rik 4930448E22Rik ## Pheta2 Pheta2 ## Gm29863 Gm29863 ## Fam89b1 Fam89b ## Spsb21 Spsb2 ## Adnp Adnp ## Gm54256 Gm54256 ## Gm15478 Gm15478 ## Tmeff1 Tmeff1 ## Prcc Prcc ## 9030612E09Rik 9030612E09Rik ## Zfp783 Zfp783 ## Ggnbp1 Ggnbp1 ## Snhg14 Snhg14 ## Gm35374 Gm35374 ## Mtnr1a Mtnr1a ## Zfp1006 Zfp1006 ## Wdr31 Wdr31 ## Tub Tub ## Agbl5 Agbl5 ## Iqch Iqch ## Gm52500 Gm52500 ## Klhl25 Klhl25 ## Stambpl1 Stambpl1 ## Gm19744 Gm19744 ## Gm31434 Gm31434 ## Vrk2 Vrk2 ## Dcun1d3 Dcun1d3 ## Metap21 Metap2 ## Ncoa5 Ncoa5 ## Acyp2 Acyp2 ## Nr1h3 Nr1h3 ## Pet117 Pet117 ## Ddx47 Ddx47 ## 2700049A03Rik 2700049A03Rik ## Gm47340 Gm47340 ## Srf1 Srf ## Trpt1 Trpt1 ## Eme2 Eme2 ## Fitm2 Fitm2 ## Cluap1 Cluap1 ## Urm1 Urm1 ## Mib2 Mib2 ## Rab40b Rab40b ## Mterf1b Mterf1b ## Arf2 Arf2 ## Pinlyp Pinlyp ## Nceh1 Nceh1 ## Lrrc49 Lrrc49 ## Gm3363 Gm3363 ## Ndufaf1 Ndufaf1 ## Emc11 Emc1 ## Sav1 Sav1 ## Cbr3 Cbr3 ## Vps51 Vps51 ## Wdr93 Wdr93 ## Tekt41 Tekt4 ## Zdhhc6 Zdhhc6 ## F830010H11Rik F830010H11Rik ## Zfp790 Zfp790 ## Fdxr Fdxr ## Pcdhgb2 Pcdhgb2 ## Fam151b Fam151b ## Rgs6 Rgs6 ## Gm11723 Gm11723 ## Guca1a Guca1a ## Mccc1 Mccc1 ## Zfp346 Zfp346 ## Gm2885 Gm2885 ## Usb1 Usb1 ## Inpp5b Inpp5b ## Dnttip21 Dnttip2 ## Snx16 Snx16 ## Pofut1 Pofut1 ## 1110002L01Rik 1110002L01Rik ## Gm41774 Gm41774 ## Ssh1 Ssh1 ## Zdhhc4 Zdhhc4 ## Nol11 Nol11 ## Gatb Gatb ## Wdtc11 Wdtc1 ## Gm53858 Gm53858 ## Ogfod1 Ogfod1 ## Phldb3 Phldb3 ## Gpsm1 Gpsm1 ## Erich5 Erich5 ## Wdr44 Wdr44 ## Fmnl1 Fmnl1 ## Mrtfa Mrtfa ## Lrrc51 Lrrc51 ## Mbip Mbip ## Twf21 Twf2 ## AW554918 AW554918 ## Fbh11 Fbh1 ## Gm35315 Gm35315 ## Gm57533 Gm57533 ## Gm5785 Gm5785 ## C330016L05Rik C330016L05Rik ## Gm16740 Gm16740 ## C030006K11Rik C030006K11Rik ## Mtmr12 Mtmr12 ## 4930458D05Rik 4930458D05Rik ## Nckap5l Nckap5l ## 1700088E04Rik 1700088E04Rik ## Brat1 Brat1 ## Abraxas2 Abraxas2 ## Zfp629 Zfp629 ## Lrig2 Lrig2 ## Tysnd1 Tysnd1 ## Vav21 Vav2 ## Scly Scly ## Sh2b3 Sh2b3 ## Polr1e Polr1e ## Nme7 Nme7 ## Abca5 Abca5 ## Gcfc2 Gcfc2 ## Pax41 Pax4 ## Znhit6 Znhit6 ## Syngr1 Syngr1 ## Tmem198b Tmem198b ## Gm14023 Gm14023 ## Zfp229 Zfp229 ## Slc6a4 Slc6a4 ## Uxt1 Uxt ## Gm20346 Gm20346 ## Rhno1 Rhno1 ## Zfp768 Zfp768 ## Crppa Crppa ## Riok2 Riok2 ## Timp2 Timp2 ## Otub2 Otub2 ## Agl Agl ## Rap1b1 Rap1b ## Sema4c Sema4c ## Zfp526 Zfp526 ## Gm154171 Gm15417 ## Kbtbd13 Kbtbd13 ## Klhl8 Klhl8 ## Homez Homez ## Rbm3os Rbm3os ## Mtr Mtr ## Sephs1 Sephs1 ## Trib3 Trib3 ## Atg71 Atg7 ## Dhrs13os Dhrs13os ## Zfp599 Zfp599 ## Phc21 Phc2 ## Bcl2l13 Bcl2l13 ## Nudt8 Nudt8 ## Tmem199 Tmem199 ## Mycl Mycl ## Osbpl7 Osbpl7 ## Fcf11 Fcf1 ## Alkbh3 Alkbh3 ## Zfp46 Zfp46 ## Paxip11 Paxip1 ## Rpp30 Rpp30 ## Hcn2 Hcn2 ## Msl3l21 Msl3l2 ## Pdp1 Pdp1 ## Ripk2 Ripk2 ## Ppp1r3e Ppp1r3e ## Ghdc Ghdc ## Arl16 Arl16 ## Zfp940 Zfp940 ## Gm31834 Gm31834 ## Phtf1 Phtf1 ## Gm20939 Gm20939 ## Slc26a10 Slc26a10 ## 2510003B16Rik 2510003B16Rik ## Gm41776 Gm41776 ## Padi1 Padi1 ## 5330438D12Rik 5330438D12Rik ## Ndufaf5 Ndufaf5 ## Gm29938 Gm29938 ## Rab28 Rab28 ## Zfp524 Zfp524 ## St3gal1 St3gal1 ## Itfg21 Itfg2 ## Rbbp9 Rbbp9 ## Gm461181 Gm46118 ## Dpysl5 Dpysl5 ## Zfp1007 Zfp1007 ## Exoc3l Exoc3l ## Gm40218 Gm40218 ## Gm54308 Gm54308 ## Rilpl1 Rilpl1 ## 1700086P04Rik 1700086P04Rik ## Gon7 Gon7 ## Zfp532 Zfp532 ## Lyrm7 Lyrm7 ## Wnt2b Wnt2b ## Gm41638 Gm41638 ## Ubxn81 Ubxn8 ## AI429214 AI429214 ## Abcb11 Abcb11 ## Armcx6 Armcx6 ## Shisa51 Shisa5 ## Gm53993 Gm53993 ## Isca11 Isca1 ## Gm16023 Gm16023 ## Sass6 Sass6 ## Gm54192 Gm54192 ## Ank1 Ank1 ## Gm53893 Gm53893 ## Ap1s2 Ap1s2 ## Gm53774 Gm53774 ## Gm35946 Gm35946 ## Klhl21 Klhl2 ## Tbx3os11 Tbx3os1 ## Sirt4 Sirt4 ## Uba6 Uba6 ## Zfp169 Zfp169 ## Ift56 Ift56 ## G630018N14Rik G630018N14Rik ## Fto Fto ## Mogat1 Mogat1 ## Smg8 Smg8 ## Vps45 Vps45 ## Taf5 Taf5 ## Syt3 Syt3 ## Nupr2 Nupr2 ## Sfxn4 Sfxn4 ## Tsga10 Tsga10 ## Mbtps2 Mbtps2 ## Eif3c1 Eif3c ## Rassf6 Rassf6 ## Gm11337 Gm11337 ## LOC118568635 LOC118568635 ## Gm33149 Gm33149 ## Slc4a1ap Slc4a1ap ## Ccdc62 Ccdc62 ## Larp7 Larp7 ## Lrrc61 Lrrc61 ## B3gnt7 B3gnt7 ## Lrtm2 Lrtm2 ## Gm53375 Gm53375 ## E130310I04Rik E130310I04Rik ## 9330151L19Rik 9330151L19Rik ## D030040B21Rik D030040B21Rik ## 4930487H11Rik 4930487H11Rik ## Gm19708 Gm19708 ## Gm32592 Gm32592 ## Gm41406 Gm41406 ## Actn3 Actn3 ## 9430060I03Rik 9430060I03Rik ## Gm50627 Gm50627 ## Gm39188 Gm39188 ## Gm39887 Gm39887 ## Gm47792 Gm47792 ## Gm57794 Gm57794 ## Gm46174 Gm46174 ## Gm53300 Gm53300 ## Gm16244 Gm16244 ## Gm30025 Gm30025 ## Gm46137 Gm46137 ## Adat1 Adat1 ## Hdac11 Hdac11 ## Yy2 Yy2 ## Alg31 Alg3 ## Zfp316 Zfp316 ## 5830444B04Rik 5830444B04Rik ## Irgq Irgq ## Zbtb39 Zbtb39 ## Fbxl9 Fbxl9 ## 1810062G17Rik 1810062G17Rik ## Glb1l Glb1l ## Vcpkmt Vcpkmt ## Cep20 Cep20 ## Usp46 Usp46 ## Gm53985 Gm53985 ## Gm46901 Gm46901 ## 9130019P16Rik 9130019P16Rik ## Gm39424 Gm39424 ## Ssc4d Ssc4d ## Nudt6 Nudt6 ## Gm30365 Gm30365 ## Zfp946 Zfp946 ## Gm30934 Gm30934 ## Bloc1s4 Bloc1s4 ## Armcx5 Armcx5 ## Rfc3 Rfc3 ## Lig4 Lig4 ## Bbs10 Bbs10 ## Pex141 Pex14 ## Wdr18 Wdr18 ## Rexo5 Rexo5 ## Ccdc166 Ccdc166 ## Zfp97 Zfp97 ## Zfp108 Zfp108 ## Zfp27 Zfp27 ## Sh2d3c Sh2d3c ## Cdk17 Cdk17 ## Nts Nts ## Telo2 Telo2 ## Mt3 Mt3 ## Arl14ep Arl14ep ## Zfp958 Zfp958 ## Mre11a Mre11a ## Abcb4 Abcb4 ## Zfp984 Zfp984 ## Cmss1 Cmss1 ## Elk1 Elk1 ## 3110045C21Rik 3110045C21Rik ## Gm32856 Gm32856 ## Insl5 Insl5 ## Arsk Arsk ## Matn2 Matn2 ## Elovl2 Elovl2 ## Setd4 Setd4 ## Kif9 Kif9 ## Gm41466 Gm41466 ## Gm54057 Gm54057 ## Cyp39a1 Cyp39a1 ## Pdxp Pdxp ## Trpc3 Trpc3 ## Tmlhe Tmlhe ## Rbm12b1 Rbm12b1 ## Mrpl49 Mrpl49 ## C030034I22Rik C030034I22Rik ## Tubg2 Tubg2 ## A930006K02Rik A930006K02Rik ## Ankrd49 Ankrd49 ## Zfp719 Zfp719 ## Gm36684 Gm36684 ## Xrcc2 Xrcc2 ## Efnb2 Efnb2 ## Ints14 Ints14 ## Gm10244 Gm10244 ## Zfp84 Zfp84 ## Styx Styx ## Rtl8c Rtl8c ## Cngb1 Cngb1 ## Gm46043 Gm46043 ## Pde8b Pde8b ## AU041133 AU041133 ## Sv2a Sv2a ## Zfp433 Zfp433 ## Lrrc40 Lrrc40 ## Tnfsf13b Tnfsf13b ## Crtc1 Crtc1 ## Hdgfl2 Hdgfl2 ## Gm13524 Gm13524 ## Iqcg Iqcg ## Rab26os Rab26os ## Trim5 Trim5 ## Gm51817 Gm51817 ## Slc10a3 Slc10a3 ## Gm34045 Gm34045 ## Gm40208 Gm40208 ## Top1mt Top1mt ## Clasp2 Clasp2 ## Lmo2 Lmo2 ## Wrap53 Wrap53 ## Fkrp1 Fkrp ## Gm14325 Gm14325 ## Cdyl2 Cdyl2 ## Prmt9 Prmt9 ## Klf16 Klf16 ## Rwdd3 Rwdd3 ## Ap3m2 Ap3m2 ## Gm31364 Gm31364 ## Poc5 Poc5 ## Zfp354b Zfp354b ## Bckdhb Bckdhb ## Xab2 Xab2 ## Gm10463 Gm10463 ## Nop9 Nop9 ## Gm38673 Gm38673 ## Acp6 Acp6 ## Ubxn2b Ubxn2b ## Slc26a11 Slc26a11 ## Adamts15 Adamts15 ## Gm51801 Gm51801 ## Adrb1 Adrb1 ## 6330418K02Rik 6330418K02Rik ## Scfd2 Scfd2 ## Mocs3 Mocs3 ## Tusc1 Tusc1 ## Wdr91 Wdr91 ## Gm36117 Gm36117 ## Riox1 Riox1 ## Faxc Faxc ## Gm50602 Gm50602 ## Gm15483 Gm15483 ## Oxnad1 Oxnad1 ## Numbl Numbl ## Rpl37rt Rpl37rt ## BC037039 BC037039 ## Tmem65 Tmem65 ## Cgnl1 Cgnl1 ## Dynlt1b Dynlt1b ## Abraxas1 Abraxas1 ## Rgs12 Rgs12 ## Mcub Mcub ## Meak7 Meak7 ## Pcdhb15 Pcdhb15 ## Spin4 Spin4 ## Slc35b4 Slc35b4 ## Gm36359 Gm36359 ## Stam Stam ## Gm40733 Gm40733 ## Gm14393 Gm14393 ## Gm32313 Gm32313 ## Hykk Hykk ## Gm38486 Gm38486 ## Spire2 Spire2 ## 1700018L02Rik 1700018L02Rik ## Asb1 Asb1 ## Gm36449 Gm36449 ## Hspa13 Hspa13 ## Rpgrip1 Rpgrip1 ## Gm4285 Gm4285 ## Casp12 Casp12 ## Cyb561d2 Cyb561d2 ## Alg1 Alg1 ## Gm36856 Gm36856 ## B4galt71 B4galt7 ## Stamos Stamos ## Pigb Pigb ## Gm36447 Gm36447 ## Gm54309 Gm54309 ## 9030624G23Rik 9030624G23Rik ## 4930511A08Rik 4930511A08Rik ## Polr3f Polr3f ## Gm13572 Gm13572 ## Gm30074 Gm30074 ## Banf2 Banf2 ## Gm53451 Gm53451 ## Cetn4 Cetn4 ## Oas1c Oas1c ## Gm54038 Gm54038 ## Erc2 Erc2 ## Gm40371 Gm40371 ## Rab8b Rab8b ## Gm46933 Gm46933 ## Zkscan4 Zkscan4 ## Cab39l Cab39l ## Wdr55 Wdr55 ## Mmrn2 Mmrn2 ## Wdr35 Wdr35 ## Gm38604 Gm38604 ## Shroom4 Shroom4 ## Gm40572 Gm40572 ## Flrt3 Flrt3 ## Mtg2 Mtg2 ## Gm34362 Gm34362 ## Asb3 Asb3 ## Lrrc28 Lrrc28 ``` --- # Find Markers And we can then visualise this using the VlnPlot function. ``` r VlnPlot(Nd1T.obj.filt, features = "Pzp") ``` <!-- -->