class: center, middle, inverse, title-slide .title[ # Reproducible Reports<br /> <html><br /> <br /> <hr color='#EB811B' size=1px width=796px><br /> </html> ] .author[ ### Rockefeller University, Bioinformatics Resource Centre ] .date[ ### <a href="https://rockefelleruniversity.github.io/RU_reproducibleR/" class="uri">https://rockefelleruniversity.github.io/RU_reproducibleR/</a> ] --- ## Overview - [Set up](https://rockefelleruniversity.github.io/RU_introtoR_abridged/presentations/singlepage/introToR_Session1.html#set-up) - [Background to R](https://rockefelleruniversity.github.io/RU_introtoR_abridged/presentations/singlepage/introToR_Session1.html#background-to-r) - [Data types in R](https://rockefelleruniversity.github.io/RU_introtoR_abridged/presentations/singlepage/introToR_Session1.html#data_types_in_r) --- class: inverse, center, middle # Set Up <html><div style='float:left'></div><hr color='#EB811B' size=1px width=720px></html> --- ## Set Up All prerequisites, links to material and slides for this course can be found on github. * [Reproducible_R](https://rockefelleruniversity.github.io/Reproducible_R/) Or can be downloaded as a zip archive from here. * [Download zip](https://github.com/rockefelleruniversity/Reproducible_R/zipball/master) --- ## Course materials Once the zip file in unarchived. All presentations as HTML slides and pages, their R code and HTML practical sheets will be available in the directories underneath. * **presentations/slides/** Presentations as an HTML slide show. * **presentations/singlepage/** Presentations as an HTML single page. * **presentations/r_code/** R code in presentations. * **exercises/** Practicals as HTML pages. * **answers/** Practicals with answers as HTML pages and R code solutions. --- ## Set the Working directory Before running any of the code in the practicals or slides we need to set the working directory to the folder we unarchived. You may navigate to the unarchived Reproducible_R folder in the Rstudio menu. **Session -> Set Working Directory -> Choose Directory** or in the console. ``` r setwd("/PathToMyDownload/Reproducible_R-master/r_course") # e.g. setwd('~/Downloads/Reproducible_R-master/r_course') ``` --- class: inverse, center, middle # Reproducible Research <html><div style='float:left'></div><hr color='#EB811B' size=1px width=720px></html> --- --- ## Reproducible Research >"Let us change our traditional attitude to the construction of programs: Instead of imagining that our main task is to instruct a computer what to do, let us concentrate rather on explaining to humans what we want the computer to do." -- Donald E. Knuth, Literate Programming, 1984 --- ## Reproducible Research in R Sometime in the future, I, or someone else, will need to understand what analysis I did here. There is a growing push to ensure all research is open and reproducible. New NIH guidelines are going to require plans for preservation and sharing of data, which includes your code. Using RStudio to make reproducible documents is very easy, so why not? --- ## A very quick reproducible document in R - Find your R script of interest. - Add comments with # to describe what you are doing. - Add the sessionInfo() function to the last line. - Click the "Compile Notebook function" -> Select HTML document as output format. ``` r # Generate some random numbers and plot them myRandNumbers <- rnorm(100, 10, 2) plot(myRandNumbers) sessionInfo() ``` --- class: inverse, center, middle # R Markdown <html><div style='float:left'></div><hr color='#EB811B' size=1px width=720px></html> --- ## The Gold Standard When we do some analysis the ideal situation is the preservation od ALL aspects of the analysis in a single document. Rmarkdown can help with this. [Here](scripts/markdownExample.html) is an example of a report generated from Rmd. The Rmd from which this is made is in the scripts directory. *scripts/markdownExample.html* *scripts/markdownExample.Rmd* --- ## R Markdown R Markdown is built on Markdown. Github and Sourceforge make use of Markdown syntax in their Readme files and renders these in their web pages. It is also used by other notebook tools like Jupyter. https://github.com/github/markup/blob/master/README.md --- ## Markdown syntax Markdown uses simple syntax to control text output. This allows for the inclusion of font styles, text structures, images and code chunks. --- ## R Markdown Structure There are 3 main parts 1. Header - YAML format 2. Body - Markdown 3. Code Chunks - R, python, shell etc. --- class: inverse, center, middle # R Markdown Header <html><div style='float:left'></div><hr color='#EB811B' size=1px width=720px></html> --- ## R Markdown Header In R Markdown the options for document processing are stored in YAML format at the top of the document. Most of this is automatically generated when you open a new Rmd. ``` default --- title: "My New Analysis" author: "Tom Carroll" date: "19th October 2022" output: html_document --- ``` --- ## Controlling output type The **output** YAML option specifies the document type to be produced. THere are several options. ``` default --- output: html_document --- ``` ``` default --- output: pdf_document --- ``` ``` default --- output: ioslides_presentation --- ``` --- ## Figure options in YAML Global default options can also be set in the YAML. For example figure sizes can be set within the YAML metadata. ``` default --- output: html_document: fig_width: 7 fig_height: 6 --- ``` --- ## Adding styles Styles for HTML can be applied using the **theme** option and syntax highlighting styles control by the **highlight** option. ``` default --- output: html_document: theme: journal highlight: espresso --- ``` For a full list of theme options see - http://rmarkdown.rstudio.com/html_document_format.html --- class: inverse, center, middle # R Markdown Body <html><div style='float:left'></div><hr color='#EB811B' size=1px width=720px></html> --- ## R Markdown Body The free form text and annotation is set in Markdown. This written as plain text and has a specific set of formatting rules. For example it ignores new lines. To include a new line in markdown, you need to end the previous line with two spaces. ``` default This is my first line. # the comment here shows the line end This would be a new line. This wouldn't be a new line. ``` This is my first line. This would be a new line. This wouldn't be a new line. --- ## Font emphasis Emphasis can be added to text in markdown documents using either the **_** or __*__ ``` default Italics = _Italics_ or *Italics* Bold = __Bold__ or **Bold** ``` _Italics_ __Bold__ --- ## Including images Figures or external images can be used in markdown documents. Files may be local or accessible from http URL. ``` default   ``` --- ## HTML links HTML links can be included in Markdown documents either by simply including address in text or by using **[]** for the phrase to add link to, followed the link in **()** ``` default https://rockefelleruniversity.github.io/ [Github site](https://rockefelleruniversity.github.io/) ``` https://rockefelleruniversity.github.io/ [Github site](https://rockefelleruniversity.github.io/) --- ## Creating headers Section headers can be added to Markdown documents. Headers follow the same conventions as used in HTML markup and can implemented at multiple levels of size. Section headers in Markdown are created by using the **#** symbol ``` default # Top level section ## Middle level section ### Bottom level section ``` ### Rendered Bottom level section --- ## Lists Lists can be created in Markdown using the __-__ symbol. Nested lists be specified with **+** symbol. ``` default - First item - Second item + Second item A + Second item B ``` My list rendered: - First item - Second item + Second item A + Second item B --- class: inverse, center, middle # R Markdown Code Chunks <html><div style='float:left'></div><hr color='#EB811B' size=1px width=720px></html> --- ## Code chunks In RMarkdown, text may be highlighted as if code by placing the text between apostrophes in triplicate: '''. The engine used to evaluate the code is in curly brackets, in this case R. ```` default ```{r} hist(rnorm(100)) ``` ```` This is what the code chunk renders to in your report: ``` r hist(rnorm(100)) ``` <!-- --> --- ## Code chunk options Many other options can be supplied to an individual code chunk R can produce a lot of output not related to your results. To control whether messages and warnings are reported in the rendered document we can specify the **message** and **warning** arguments. Loading libraries in R Markdown is often somewhere you would specify these as FALSE. ```` default ```{r,warning=F,message=F} library(ggplot2) ``` ```` --- ## Code chunk options There are many chunk control options * eval - Run the code? * echo - Include code in final report? * tidy - tidy up the code? * cache - save a cache * fig.height/fig.width - size of plot made by code * fig.path and dev - save plots to specified path in specified format --- class: inverse, center, middle # Other R Markdown tricks <html><div style='float:left'></div><hr color='#EB811B' size=1px width=720px></html> --- ## Inserting tables The results of printing data frames or matrices in the console aren't neat. We can insert tables into R Markdown by using the knitr function **kable()**. ``` r temp <- rnorm(3) temp2 <- rnorm(3) dfExample <- data.frame(temp, temp2) kable(dfExample) ``` | temp| temp2| |---------:|----------:| | 0.3183553| -0.4989068| | 0.6474253| -0.7809624| | 0.5844188| 1.8188830| --- ## Evaluating code in the body Most of your code will be in code chunks. But it may be useful to report the results of R within the block of Markdown. This can be done adding the code to evalulate within **'r '** ``` default Here is some freeform _markdown_ and the first result from an rnorm call is 'r rnorm(3)[1]', followed by some more free form text. ``` Here is some freeform _markdown_ and the first result from an rnorm call is 1.708487, followed by some more free form text. --- class: inverse, center, middle # Interactivity <html><div style='float:left'></div><hr color='#EB811B' size=1px width=720px></html> --- ## Interactivity As we are summarizing data in a dynamic document, we can make data exporation easier by creating interactive versions of plots and tables. ``` ## Loading required package: ggplot2 ``` ``` ## ## Attaching package: 'plotly' ``` ``` ## The following object is masked from 'package:ggplot2': ## ## last_plot ``` ``` ## The following object is masked from 'package:IRanges': ## ## slice ``` ``` ## The following object is masked from 'package:S4Vectors': ## ## rename ``` ``` ## The following object is masked from 'package:stats': ## ## filter ``` ``` ## The following object is masked from 'package:graphics': ## ## layout ``` <div class="plotly html-widget html-fill-item" id="htmlwidget-0520b360a2c618a1df74" style="width:504px;height:504px;"></div> <script type="application/json" data-for="htmlwidget-0520b360a2c618a1df74">{"x":{"data":[{"x":[0.63672798475643777],"y":[0.61639057522774621],"text":"PC1: 0.6367280<br />PC2: 0.6163906<br />Time: 0hr<br />Rep: 1","type":"scatter","mode":"markers","marker":{"autocolorscale":false,"color":"rgba(248,118,109,1)","opacity":1,"size":5.6692913385826778,"symbol":"circle","line":{"width":1.8897637795275593,"color":"rgba(248,118,109,1)"}},"hoveron":"points","name":"(0hr,1)","legendgroup":"(0hr,1)","showlegend":true,"xaxis":"x","yaxis":"y","hoverinfo":"text","frame":null},{"x":[0.60906629999999995],"y":[0.53843799999999997],"text":"PC1: 0.6090663<br />PC2: 0.5384380<br />Time: 0hr<br />Rep: 2","type":"scatter","mode":"markers","marker":{"autocolorscale":false,"color":"rgba(248,118,109,1)","opacity":1,"size":5.6692913385826778,"symbol":"triangle-up","line":{"width":1.8897637795275593,"color":"rgba(248,118,109,1)"}},"hoveron":"points","name":"(0hr,2)","legendgroup":"(0hr,2)","showlegend":true,"xaxis":"x","yaxis":"y","hoverinfo":"text","frame":null},{"x":[0.4596307],"y":[0.54244119999999996],"text":"PC1: 0.4596307<br />PC2: 0.5424412<br />Time: 2hr<br />Rep: 1","type":"scatter","mode":"markers","marker":{"autocolorscale":false,"color":"rgba(0,186,56,1)","opacity":1,"size":5.6692913385826778,"symbol":"circle","line":{"width":1.8897637795275593,"color":"rgba(0,186,56,1)"}},"hoveron":"points","name":"(2hr,1)","legendgroup":"(2hr,1)","showlegend":true,"xaxis":"x","yaxis":"y","hoverinfo":"text","frame":null},{"x":[0.41904629999999998],"y":[0.48124299999999998],"text":"PC1: 0.4190463<br />PC2: 0.4812430<br />Time: 2hr<br />Rep: 2","type":"scatter","mode":"markers","marker":{"autocolorscale":false,"color":"rgba(0,186,56,1)","opacity":1,"size":5.6692913385826778,"symbol":"triangle-up","line":{"width":1.8897637795275593,"color":"rgba(0,186,56,1)"}},"hoveron":"points","name":"(2hr,2)","legendgroup":"(2hr,2)","showlegend":true,"xaxis":"x","yaxis":"y","hoverinfo":"text","frame":null},{"x":[0.32207960000000002],"y":[0.54347190000000001],"text":"PC1: 0.3220796<br />PC2: 0.5434719<br />Time: 12hr<br />Rep: 1","type":"scatter","mode":"markers","marker":{"autocolorscale":false,"color":"rgba(97,156,255,1)","opacity":1,"size":5.6692913385826778,"symbol":"circle","line":{"width":1.8897637795275593,"color":"rgba(97,156,255,1)"}},"hoveron":"points","name":"(12hr,1)","legendgroup":"(12hr,1)","showlegend":true,"xaxis":"x","yaxis":"y","hoverinfo":"text","frame":null},{"x":[0.31305909999999998],"y":[0.44264419999999999],"text":"PC1: 0.3130591<br />PC2: 0.4426442<br />Time: 12hr<br />Rep: 2","type":"scatter","mode":"markers","marker":{"autocolorscale":false,"color":"rgba(97,156,255,1)","opacity":1,"size":5.6692913385826778,"symbol":"triangle-up","line":{"width":1.8897637795275593,"color":"rgba(97,156,255,1)"}},"hoveron":"points","name":"(12hr,2)","legendgroup":"(12hr,2)","showlegend":true,"xaxis":"x","yaxis":"y","hoverinfo":"text","frame":null}],"layout":{"margin":{"t":23.305936073059364,"r":7.3059360730593621,"b":37.260273972602747,"l":48.949771689497723},"plot_bgcolor":"rgba(235,235,235,1)","paper_bgcolor":"rgba(255,255,255,1)","font":{"color":"rgba(0,0,0,1)","family":"","size":14.611872146118724},"xaxis":{"domain":[0,1],"automargin":true,"type":"linear","autorange":false,"range":[0.29687565576217811,0.65291142899425969],"tickmode":"array","ticktext":["0.3","0.4","0.5","0.6"],"tickvals":[0.30000000000000004,0.40000000000000002,0.5,0.60000000000000009],"categoryorder":"array","categoryarray":["0.3","0.4","0.5","0.6"],"nticks":null,"ticks":"outside","tickcolor":"rgba(51,51,51,1)","ticklen":3.6529680365296811,"tickwidth":0.66417600664176002,"showticklabels":true,"tickfont":{"color":"rgba(77,77,77,1)","family":"","size":11.68949771689498},"tickangle":-0,"showline":false,"linecolor":null,"linewidth":0,"showgrid":true,"gridcolor":"rgba(255,255,255,1)","gridwidth":0.66417600664176002,"zeroline":false,"anchor":"y","title":{"text":"PC1","font":{"color":"rgba(0,0,0,1)","family":"","size":14.611872146118724}},"hoverformat":".2f"},"yaxis":{"domain":[0,1],"automargin":true,"type":"linear","autorange":false,"range":[0.4339568812386127,0.62507789398913349],"tickmode":"array","ticktext":["0.45","0.50","0.55","0.60"],"tickvals":[0.45000000000000001,0.5,0.55000000000000004,0.60000000000000009],"categoryorder":"array","categoryarray":["0.45","0.50","0.55","0.60"],"nticks":null,"ticks":"outside","tickcolor":"rgba(51,51,51,1)","ticklen":3.6529680365296811,"tickwidth":0.66417600664176002,"showticklabels":true,"tickfont":{"color":"rgba(77,77,77,1)","family":"","size":11.68949771689498},"tickangle":-0,"showline":false,"linecolor":null,"linewidth":0,"showgrid":true,"gridcolor":"rgba(255,255,255,1)","gridwidth":0.66417600664176002,"zeroline":false,"anchor":"x","title":{"text":"PC2","font":{"color":"rgba(0,0,0,1)","family":"","size":14.611872146118724}},"hoverformat":".2f"},"shapes":[{"type":"rect","fillcolor":null,"line":{"color":null,"width":0,"linetype":[]},"yref":"paper","xref":"paper","x0":0,"x1":1,"y0":0,"y1":1}],"showlegend":true,"legend":{"bgcolor":"rgba(255,255,255,1)","bordercolor":"transparent","borderwidth":1.8897637795275593,"font":{"color":"rgba(0,0,0,1)","family":"","size":11.68949771689498},"title":{"text":"Time<br />Rep","font":{"color":"rgba(0,0,0,1)","family":"","size":14.611872146118724}}},"hovermode":"closest","barmode":"relative"},"config":{"doubleClick":"reset","modeBarButtonsToAdd":["hoverclosest","hovercompare"],"showSendToCloud":false},"source":"A","attrs":{"169e23f85197":{"x":{},"y":{},"colour":{},"shape":{},"type":"scatter"}},"cur_data":"169e23f85197","visdat":{"169e23f85197":["function (y) ","x"]},"highlight":{"on":"plotly_click","persistent":false,"dynamic":false,"selectize":false,"opacityDim":0.20000000000000001,"selected":{"opacity":1},"debounce":0},"shinyEvents":["plotly_hover","plotly_click","plotly_selected","plotly_relayout","plotly_brushed","plotly_brushing","plotly_clickannotation","plotly_doubleclick","plotly_deselect","plotly_afterplot","plotly_sunburstclick"],"base_url":"https://plot.ly"},"evals":[],"jsHooks":[]}</script> --- ## Plotly To do this we use the *plotly* package. Lets load in the package and the plot we want to make interactive. We have an R object that contains a ggplot2 object in our *data* directory. ``` r library(plotly) load("data/pcPlot.RData") pcPlot ``` <!-- --> --- ## Plotly labels The *ggplotly* function will wrap our ggplot object and add interactivity so we can hover over the points. ``` r ggplotly(pcPlot) ``` <div class="plotly html-widget html-fill-item" id="htmlwidget-a713d866601547724a1c" style="width:504px;height:504px;"></div> <script type="application/json" data-for="htmlwidget-a713d866601547724a1c">{"x":{"data":[{"x":[0.63672798475643777],"y":[0.61639057522774621],"text":"PC1: 0.6367280<br />PC2: 0.6163906<br />Time: 0hr<br />Rep: 1","type":"scatter","mode":"markers","marker":{"autocolorscale":false,"color":"rgba(248,118,109,1)","opacity":1,"size":5.6692913385826778,"symbol":"circle","line":{"width":1.8897637795275593,"color":"rgba(248,118,109,1)"}},"hoveron":"points","name":"(0hr,1)","legendgroup":"(0hr,1)","showlegend":true,"xaxis":"x","yaxis":"y","hoverinfo":"text","frame":null},{"x":[0.60906629999999995],"y":[0.53843799999999997],"text":"PC1: 0.6090663<br />PC2: 0.5384380<br />Time: 0hr<br />Rep: 2","type":"scatter","mode":"markers","marker":{"autocolorscale":false,"color":"rgba(248,118,109,1)","opacity":1,"size":5.6692913385826778,"symbol":"triangle-up","line":{"width":1.8897637795275593,"color":"rgba(248,118,109,1)"}},"hoveron":"points","name":"(0hr,2)","legendgroup":"(0hr,2)","showlegend":true,"xaxis":"x","yaxis":"y","hoverinfo":"text","frame":null},{"x":[0.4596307],"y":[0.54244119999999996],"text":"PC1: 0.4596307<br />PC2: 0.5424412<br />Time: 2hr<br />Rep: 1","type":"scatter","mode":"markers","marker":{"autocolorscale":false,"color":"rgba(0,186,56,1)","opacity":1,"size":5.6692913385826778,"symbol":"circle","line":{"width":1.8897637795275593,"color":"rgba(0,186,56,1)"}},"hoveron":"points","name":"(2hr,1)","legendgroup":"(2hr,1)","showlegend":true,"xaxis":"x","yaxis":"y","hoverinfo":"text","frame":null},{"x":[0.41904629999999998],"y":[0.48124299999999998],"text":"PC1: 0.4190463<br />PC2: 0.4812430<br />Time: 2hr<br />Rep: 2","type":"scatter","mode":"markers","marker":{"autocolorscale":false,"color":"rgba(0,186,56,1)","opacity":1,"size":5.6692913385826778,"symbol":"triangle-up","line":{"width":1.8897637795275593,"color":"rgba(0,186,56,1)"}},"hoveron":"points","name":"(2hr,2)","legendgroup":"(2hr,2)","showlegend":true,"xaxis":"x","yaxis":"y","hoverinfo":"text","frame":null},{"x":[0.32207960000000002],"y":[0.54347190000000001],"text":"PC1: 0.3220796<br />PC2: 0.5434719<br />Time: 12hr<br />Rep: 1","type":"scatter","mode":"markers","marker":{"autocolorscale":false,"color":"rgba(97,156,255,1)","opacity":1,"size":5.6692913385826778,"symbol":"circle","line":{"width":1.8897637795275593,"color":"rgba(97,156,255,1)"}},"hoveron":"points","name":"(12hr,1)","legendgroup":"(12hr,1)","showlegend":true,"xaxis":"x","yaxis":"y","hoverinfo":"text","frame":null},{"x":[0.31305909999999998],"y":[0.44264419999999999],"text":"PC1: 0.3130591<br />PC2: 0.4426442<br />Time: 12hr<br />Rep: 2","type":"scatter","mode":"markers","marker":{"autocolorscale":false,"color":"rgba(97,156,255,1)","opacity":1,"size":5.6692913385826778,"symbol":"triangle-up","line":{"width":1.8897637795275593,"color":"rgba(97,156,255,1)"}},"hoveron":"points","name":"(12hr,2)","legendgroup":"(12hr,2)","showlegend":true,"xaxis":"x","yaxis":"y","hoverinfo":"text","frame":null}],"layout":{"margin":{"t":23.305936073059364,"r":7.3059360730593621,"b":37.260273972602747,"l":48.949771689497723},"plot_bgcolor":"rgba(235,235,235,1)","paper_bgcolor":"rgba(255,255,255,1)","font":{"color":"rgba(0,0,0,1)","family":"","size":14.611872146118724},"xaxis":{"domain":[0,1],"automargin":true,"type":"linear","autorange":false,"range":[0.29687565576217811,0.65291142899425969],"tickmode":"array","ticktext":["0.3","0.4","0.5","0.6"],"tickvals":[0.30000000000000004,0.40000000000000002,0.5,0.60000000000000009],"categoryorder":"array","categoryarray":["0.3","0.4","0.5","0.6"],"nticks":null,"ticks":"outside","tickcolor":"rgba(51,51,51,1)","ticklen":3.6529680365296811,"tickwidth":0.66417600664176002,"showticklabels":true,"tickfont":{"color":"rgba(77,77,77,1)","family":"","size":11.68949771689498},"tickangle":-0,"showline":false,"linecolor":null,"linewidth":0,"showgrid":true,"gridcolor":"rgba(255,255,255,1)","gridwidth":0.66417600664176002,"zeroline":false,"anchor":"y","title":{"text":"PC1","font":{"color":"rgba(0,0,0,1)","family":"","size":14.611872146118724}},"hoverformat":".2f"},"yaxis":{"domain":[0,1],"automargin":true,"type":"linear","autorange":false,"range":[0.4339568812386127,0.62507789398913349],"tickmode":"array","ticktext":["0.45","0.50","0.55","0.60"],"tickvals":[0.45000000000000001,0.5,0.55000000000000004,0.60000000000000009],"categoryorder":"array","categoryarray":["0.45","0.50","0.55","0.60"],"nticks":null,"ticks":"outside","tickcolor":"rgba(51,51,51,1)","ticklen":3.6529680365296811,"tickwidth":0.66417600664176002,"showticklabels":true,"tickfont":{"color":"rgba(77,77,77,1)","family":"","size":11.68949771689498},"tickangle":-0,"showline":false,"linecolor":null,"linewidth":0,"showgrid":true,"gridcolor":"rgba(255,255,255,1)","gridwidth":0.66417600664176002,"zeroline":false,"anchor":"x","title":{"text":"PC2","font":{"color":"rgba(0,0,0,1)","family":"","size":14.611872146118724}},"hoverformat":".2f"},"shapes":[{"type":"rect","fillcolor":null,"line":{"color":null,"width":0,"linetype":[]},"yref":"paper","xref":"paper","x0":0,"x1":1,"y0":0,"y1":1}],"showlegend":true,"legend":{"bgcolor":"rgba(255,255,255,1)","bordercolor":"transparent","borderwidth":1.8897637795275593,"font":{"color":"rgba(0,0,0,1)","family":"","size":11.68949771689498},"title":{"text":"Time<br />Rep","font":{"color":"rgba(0,0,0,1)","family":"","size":14.611872146118724}}},"hovermode":"closest","barmode":"relative"},"config":{"doubleClick":"reset","modeBarButtonsToAdd":["hoverclosest","hovercompare"],"showSendToCloud":false},"source":"A","attrs":{"169e4ff25b86":{"x":{},"y":{},"colour":{},"shape":{},"type":"scatter"}},"cur_data":"169e4ff25b86","visdat":{"169e4ff25b86":["function (y) ","x"]},"highlight":{"on":"plotly_click","persistent":false,"dynamic":false,"selectize":false,"opacityDim":0.20000000000000001,"selected":{"opacity":1},"debounce":0},"shinyEvents":["plotly_hover","plotly_click","plotly_selected","plotly_relayout","plotly_brushed","plotly_brushing","plotly_clickannotation","plotly_doubleclick","plotly_deselect","plotly_afterplot","plotly_sunburstclick"],"base_url":"https://plot.ly"},"evals":[],"jsHooks":[]}</script> --- ## Plotly labels The information assocaited with the plot are naturally inherited as labels. If we want to add custom labels we can specify it in our plot. ``` r ggplotly(pcPlot + geom_point(aes(label = Sample))) ``` ``` ## Warning in geom_point(aes(label = Sample)): Ignoring unknown aesthetics: label ``` <div class="plotly html-widget html-fill-item" id="htmlwidget-42602a83b02d7d9dab75" style="width:504px;height:504px;"></div> <script type="application/json" data-for="htmlwidget-42602a83b02d7d9dab75">{"x":{"data":[{"x":[0.63672798475643777],"y":[0.61639057522774621],"text":"PC1: 0.6367280<br />PC2: 0.6163906<br />Time: 0hr<br />Rep: 1","type":"scatter","mode":"markers","marker":{"autocolorscale":false,"color":"rgba(248,118,109,1)","opacity":1,"size":5.6692913385826778,"symbol":"circle","line":{"width":1.8897637795275593,"color":"rgba(248,118,109,1)"}},"hoveron":"points","name":"(0hr,1)","legendgroup":"(0hr,1)","showlegend":true,"xaxis":"x","yaxis":"y","hoverinfo":"text","frame":null},{"x":[0.60906629999999995],"y":[0.53843799999999997],"text":"PC1: 0.6090663<br />PC2: 0.5384380<br />Time: 0hr<br />Rep: 2","type":"scatter","mode":"markers","marker":{"autocolorscale":false,"color":"rgba(248,118,109,1)","opacity":1,"size":5.6692913385826778,"symbol":"triangle-up","line":{"width":1.8897637795275593,"color":"rgba(248,118,109,1)"}},"hoveron":"points","name":"(0hr,2)","legendgroup":"(0hr,2)","showlegend":true,"xaxis":"x","yaxis":"y","hoverinfo":"text","frame":null},{"x":[0.4596307],"y":[0.54244119999999996],"text":"PC1: 0.4596307<br />PC2: 0.5424412<br />Time: 2hr<br />Rep: 1","type":"scatter","mode":"markers","marker":{"autocolorscale":false,"color":"rgba(0,186,56,1)","opacity":1,"size":5.6692913385826778,"symbol":"circle","line":{"width":1.8897637795275593,"color":"rgba(0,186,56,1)"}},"hoveron":"points","name":"(2hr,1)","legendgroup":"(2hr,1)","showlegend":true,"xaxis":"x","yaxis":"y","hoverinfo":"text","frame":null},{"x":[0.41904629999999998],"y":[0.48124299999999998],"text":"PC1: 0.4190463<br />PC2: 0.4812430<br />Time: 2hr<br />Rep: 2","type":"scatter","mode":"markers","marker":{"autocolorscale":false,"color":"rgba(0,186,56,1)","opacity":1,"size":5.6692913385826778,"symbol":"triangle-up","line":{"width":1.8897637795275593,"color":"rgba(0,186,56,1)"}},"hoveron":"points","name":"(2hr,2)","legendgroup":"(2hr,2)","showlegend":true,"xaxis":"x","yaxis":"y","hoverinfo":"text","frame":null},{"x":[0.32207960000000002],"y":[0.54347190000000001],"text":"PC1: 0.3220796<br />PC2: 0.5434719<br />Time: 12hr<br />Rep: 1","type":"scatter","mode":"markers","marker":{"autocolorscale":false,"color":"rgba(97,156,255,1)","opacity":1,"size":5.6692913385826778,"symbol":"circle","line":{"width":1.8897637795275593,"color":"rgba(97,156,255,1)"}},"hoveron":"points","name":"(12hr,1)","legendgroup":"(12hr,1)","showlegend":true,"xaxis":"x","yaxis":"y","hoverinfo":"text","frame":null},{"x":[0.31305909999999998],"y":[0.44264419999999999],"text":"PC1: 0.3130591<br />PC2: 0.4426442<br />Time: 12hr<br />Rep: 2","type":"scatter","mode":"markers","marker":{"autocolorscale":false,"color":"rgba(97,156,255,1)","opacity":1,"size":5.6692913385826778,"symbol":"triangle-up","line":{"width":1.8897637795275593,"color":"rgba(97,156,255,1)"}},"hoveron":"points","name":"(12hr,2)","legendgroup":"(12hr,2)","showlegend":true,"xaxis":"x","yaxis":"y","hoverinfo":"text","frame":null},{"x":[0.63672798475643777],"y":[0.61639057522774621],"text":"PC1: 0.6367280<br />PC2: 0.6163906<br />Time: 0hr<br />Rep: 1<br />Sample: A_1","type":"scatter","mode":"markers","marker":{"autocolorscale":false,"color":"rgba(248,118,109,1)","opacity":1,"size":5.6692913385826778,"symbol":"circle","line":{"width":1.8897637795275593,"color":"rgba(248,118,109,1)"}},"hoveron":"points","name":"(0hr,1)","legendgroup":"(0hr,1)","showlegend":false,"xaxis":"x","yaxis":"y","hoverinfo":"text","frame":null},{"x":[0.60906629999999995],"y":[0.53843799999999997],"text":"PC1: 0.6090663<br />PC2: 0.5384380<br />Time: 0hr<br />Rep: 2<br />Sample: A_2","type":"scatter","mode":"markers","marker":{"autocolorscale":false,"color":"rgba(248,118,109,1)","opacity":1,"size":5.6692913385826778,"symbol":"triangle-up","line":{"width":1.8897637795275593,"color":"rgba(248,118,109,1)"}},"hoveron":"points","name":"(0hr,2)","legendgroup":"(0hr,2)","showlegend":false,"xaxis":"x","yaxis":"y","hoverinfo":"text","frame":null},{"x":[0.4596307],"y":[0.54244119999999996],"text":"PC1: 0.4596307<br />PC2: 0.5424412<br />Time: 2hr<br />Rep: 1<br />Sample: B_1","type":"scatter","mode":"markers","marker":{"autocolorscale":false,"color":"rgba(0,186,56,1)","opacity":1,"size":5.6692913385826778,"symbol":"circle","line":{"width":1.8897637795275593,"color":"rgba(0,186,56,1)"}},"hoveron":"points","name":"(2hr,1)","legendgroup":"(2hr,1)","showlegend":false,"xaxis":"x","yaxis":"y","hoverinfo":"text","frame":null},{"x":[0.41904629999999998],"y":[0.48124299999999998],"text":"PC1: 0.4190463<br />PC2: 0.4812430<br />Time: 2hr<br />Rep: 2<br />Sample: B_2","type":"scatter","mode":"markers","marker":{"autocolorscale":false,"color":"rgba(0,186,56,1)","opacity":1,"size":5.6692913385826778,"symbol":"triangle-up","line":{"width":1.8897637795275593,"color":"rgba(0,186,56,1)"}},"hoveron":"points","name":"(2hr,2)","legendgroup":"(2hr,2)","showlegend":false,"xaxis":"x","yaxis":"y","hoverinfo":"text","frame":null},{"x":[0.32207960000000002],"y":[0.54347190000000001],"text":"PC1: 0.3220796<br />PC2: 0.5434719<br />Time: 12hr<br />Rep: 1<br />Sample: C_1","type":"scatter","mode":"markers","marker":{"autocolorscale":false,"color":"rgba(97,156,255,1)","opacity":1,"size":5.6692913385826778,"symbol":"circle","line":{"width":1.8897637795275593,"color":"rgba(97,156,255,1)"}},"hoveron":"points","name":"(12hr,1)","legendgroup":"(12hr,1)","showlegend":false,"xaxis":"x","yaxis":"y","hoverinfo":"text","frame":null},{"x":[0.31305909999999998],"y":[0.44264419999999999],"text":"PC1: 0.3130591<br />PC2: 0.4426442<br />Time: 12hr<br />Rep: 2<br />Sample: C_2","type":"scatter","mode":"markers","marker":{"autocolorscale":false,"color":"rgba(97,156,255,1)","opacity":1,"size":5.6692913385826778,"symbol":"triangle-up","line":{"width":1.8897637795275593,"color":"rgba(97,156,255,1)"}},"hoveron":"points","name":"(12hr,2)","legendgroup":"(12hr,2)","showlegend":false,"xaxis":"x","yaxis":"y","hoverinfo":"text","frame":null}],"layout":{"margin":{"t":23.305936073059364,"r":7.3059360730593621,"b":37.260273972602747,"l":48.949771689497723},"plot_bgcolor":"rgba(235,235,235,1)","paper_bgcolor":"rgba(255,255,255,1)","font":{"color":"rgba(0,0,0,1)","family":"","size":14.611872146118724},"xaxis":{"domain":[0,1],"automargin":true,"type":"linear","autorange":false,"range":[0.29687565576217811,0.65291142899425969],"tickmode":"array","ticktext":["0.3","0.4","0.5","0.6"],"tickvals":[0.30000000000000004,0.40000000000000002,0.5,0.60000000000000009],"categoryorder":"array","categoryarray":["0.3","0.4","0.5","0.6"],"nticks":null,"ticks":"outside","tickcolor":"rgba(51,51,51,1)","ticklen":3.6529680365296811,"tickwidth":0.66417600664176002,"showticklabels":true,"tickfont":{"color":"rgba(77,77,77,1)","family":"","size":11.68949771689498},"tickangle":-0,"showline":false,"linecolor":null,"linewidth":0,"showgrid":true,"gridcolor":"rgba(255,255,255,1)","gridwidth":0.66417600664176002,"zeroline":false,"anchor":"y","title":{"text":"PC1","font":{"color":"rgba(0,0,0,1)","family":"","size":14.611872146118724}},"hoverformat":".2f"},"yaxis":{"domain":[0,1],"automargin":true,"type":"linear","autorange":false,"range":[0.4339568812386127,0.62507789398913349],"tickmode":"array","ticktext":["0.45","0.50","0.55","0.60"],"tickvals":[0.45000000000000001,0.5,0.55000000000000004,0.60000000000000009],"categoryorder":"array","categoryarray":["0.45","0.50","0.55","0.60"],"nticks":null,"ticks":"outside","tickcolor":"rgba(51,51,51,1)","ticklen":3.6529680365296811,"tickwidth":0.66417600664176002,"showticklabels":true,"tickfont":{"color":"rgba(77,77,77,1)","family":"","size":11.68949771689498},"tickangle":-0,"showline":false,"linecolor":null,"linewidth":0,"showgrid":true,"gridcolor":"rgba(255,255,255,1)","gridwidth":0.66417600664176002,"zeroline":false,"anchor":"x","title":{"text":"PC2","font":{"color":"rgba(0,0,0,1)","family":"","size":14.611872146118724}},"hoverformat":".2f"},"shapes":[{"type":"rect","fillcolor":null,"line":{"color":null,"width":0,"linetype":[]},"yref":"paper","xref":"paper","x0":0,"x1":1,"y0":0,"y1":1}],"showlegend":true,"legend":{"bgcolor":"rgba(255,255,255,1)","bordercolor":"transparent","borderwidth":1.8897637795275593,"font":{"color":"rgba(0,0,0,1)","family":"","size":11.68949771689498},"title":{"text":"Time<br />Rep","font":{"color":"rgba(0,0,0,1)","family":"","size":14.611872146118724}}},"hovermode":"closest","barmode":"relative"},"config":{"doubleClick":"reset","modeBarButtonsToAdd":["hoverclosest","hovercompare"],"showSendToCloud":false},"source":"A","attrs":{"169e6839840c":{"x":{},"y":{},"colour":{},"shape":{},"type":"scatter"},"169e14d3e376":{"x":{},"y":{},"colour":{},"shape":{},"label":{}}},"cur_data":"169e6839840c","visdat":{"169e6839840c":["function (y) ","x"],"169e14d3e376":["function (y) ","x"]},"highlight":{"on":"plotly_click","persistent":false,"dynamic":false,"selectize":false,"opacityDim":0.20000000000000001,"selected":{"opacity":1},"debounce":0},"shinyEvents":["plotly_hover","plotly_click","plotly_selected","plotly_relayout","plotly_brushed","plotly_brushing","plotly_clickannotation","plotly_doubleclick","plotly_deselect","plotly_afterplot","plotly_sunburstclick"],"base_url":"https://plot.ly"},"evals":[],"jsHooks":[]}</script> --- ## Plotly labels If we want full control over the labeling we can instead use the ggplotly function to take care of this for us by specifying when/what/where labels are shown. ``` r ggplotly(pcPlot + geom_point(aes(text = Sample)), source = "select", tooltip = c("Sample")) ``` ``` ## Warning in geom_point(aes(text = Sample)): Ignoring unknown aesthetics: text ``` <div class="plotly html-widget html-fill-item" id="htmlwidget-b9cff8e97e5a0bbfcf72" style="width:504px;height:504px;"></div> <script type="application/json" data-for="htmlwidget-b9cff8e97e5a0bbfcf72">{"x":{"data":[{"x":[0.63672798475643777],"y":[0.61639057522774621],"text":"","type":"scatter","mode":"markers","marker":{"autocolorscale":false,"color":"rgba(248,118,109,1)","opacity":1,"size":5.6692913385826778,"symbol":"circle","line":{"width":1.8897637795275593,"color":"rgba(248,118,109,1)"}},"hoveron":"points","name":"(0hr,1)","legendgroup":"(0hr,1)","showlegend":true,"xaxis":"x","yaxis":"y","hoverinfo":"text","frame":null},{"x":[0.60906629999999995],"y":[0.53843799999999997],"text":"","type":"scatter","mode":"markers","marker":{"autocolorscale":false,"color":"rgba(248,118,109,1)","opacity":1,"size":5.6692913385826778,"symbol":"triangle-up","line":{"width":1.8897637795275593,"color":"rgba(248,118,109,1)"}},"hoveron":"points","name":"(0hr,2)","legendgroup":"(0hr,2)","showlegend":true,"xaxis":"x","yaxis":"y","hoverinfo":"text","frame":null},{"x":[0.4596307],"y":[0.54244119999999996],"text":"","type":"scatter","mode":"markers","marker":{"autocolorscale":false,"color":"rgba(0,186,56,1)","opacity":1,"size":5.6692913385826778,"symbol":"circle","line":{"width":1.8897637795275593,"color":"rgba(0,186,56,1)"}},"hoveron":"points","name":"(2hr,1)","legendgroup":"(2hr,1)","showlegend":true,"xaxis":"x","yaxis":"y","hoverinfo":"text","frame":null},{"x":[0.41904629999999998],"y":[0.48124299999999998],"text":"","type":"scatter","mode":"markers","marker":{"autocolorscale":false,"color":"rgba(0,186,56,1)","opacity":1,"size":5.6692913385826778,"symbol":"triangle-up","line":{"width":1.8897637795275593,"color":"rgba(0,186,56,1)"}},"hoveron":"points","name":"(2hr,2)","legendgroup":"(2hr,2)","showlegend":true,"xaxis":"x","yaxis":"y","hoverinfo":"text","frame":null},{"x":[0.32207960000000002],"y":[0.54347190000000001],"text":"","type":"scatter","mode":"markers","marker":{"autocolorscale":false,"color":"rgba(97,156,255,1)","opacity":1,"size":5.6692913385826778,"symbol":"circle","line":{"width":1.8897637795275593,"color":"rgba(97,156,255,1)"}},"hoveron":"points","name":"(12hr,1)","legendgroup":"(12hr,1)","showlegend":true,"xaxis":"x","yaxis":"y","hoverinfo":"text","frame":null},{"x":[0.31305909999999998],"y":[0.44264419999999999],"text":"","type":"scatter","mode":"markers","marker":{"autocolorscale":false,"color":"rgba(97,156,255,1)","opacity":1,"size":5.6692913385826778,"symbol":"triangle-up","line":{"width":1.8897637795275593,"color":"rgba(97,156,255,1)"}},"hoveron":"points","name":"(12hr,2)","legendgroup":"(12hr,2)","showlegend":true,"xaxis":"x","yaxis":"y","hoverinfo":"text","frame":null},{"x":[0.63672798475643777],"y":[0.61639057522774621],"text":"A_1","type":"scatter","mode":"markers","marker":{"autocolorscale":false,"color":"rgba(248,118,109,1)","opacity":1,"size":5.6692913385826778,"symbol":"circle","line":{"width":1.8897637795275593,"color":"rgba(248,118,109,1)"}},"hoveron":"points","name":"(0hr,1)","legendgroup":"(0hr,1)","showlegend":false,"xaxis":"x","yaxis":"y","hoverinfo":"text","frame":null},{"x":[0.60906629999999995],"y":[0.53843799999999997],"text":"A_2","type":"scatter","mode":"markers","marker":{"autocolorscale":false,"color":"rgba(248,118,109,1)","opacity":1,"size":5.6692913385826778,"symbol":"triangle-up","line":{"width":1.8897637795275593,"color":"rgba(248,118,109,1)"}},"hoveron":"points","name":"(0hr,2)","legendgroup":"(0hr,2)","showlegend":false,"xaxis":"x","yaxis":"y","hoverinfo":"text","frame":null},{"x":[0.4596307],"y":[0.54244119999999996],"text":"B_1","type":"scatter","mode":"markers","marker":{"autocolorscale":false,"color":"rgba(0,186,56,1)","opacity":1,"size":5.6692913385826778,"symbol":"circle","line":{"width":1.8897637795275593,"color":"rgba(0,186,56,1)"}},"hoveron":"points","name":"(2hr,1)","legendgroup":"(2hr,1)","showlegend":false,"xaxis":"x","yaxis":"y","hoverinfo":"text","frame":null},{"x":[0.41904629999999998],"y":[0.48124299999999998],"text":"B_2","type":"scatter","mode":"markers","marker":{"autocolorscale":false,"color":"rgba(0,186,56,1)","opacity":1,"size":5.6692913385826778,"symbol":"triangle-up","line":{"width":1.8897637795275593,"color":"rgba(0,186,56,1)"}},"hoveron":"points","name":"(2hr,2)","legendgroup":"(2hr,2)","showlegend":false,"xaxis":"x","yaxis":"y","hoverinfo":"text","frame":null},{"x":[0.32207960000000002],"y":[0.54347190000000001],"text":"C_1","type":"scatter","mode":"markers","marker":{"autocolorscale":false,"color":"rgba(97,156,255,1)","opacity":1,"size":5.6692913385826778,"symbol":"circle","line":{"width":1.8897637795275593,"color":"rgba(97,156,255,1)"}},"hoveron":"points","name":"(12hr,1)","legendgroup":"(12hr,1)","showlegend":false,"xaxis":"x","yaxis":"y","hoverinfo":"text","frame":null},{"x":[0.31305909999999998],"y":[0.44264419999999999],"text":"C_2","type":"scatter","mode":"markers","marker":{"autocolorscale":false,"color":"rgba(97,156,255,1)","opacity":1,"size":5.6692913385826778,"symbol":"triangle-up","line":{"width":1.8897637795275593,"color":"rgba(97,156,255,1)"}},"hoveron":"points","name":"(12hr,2)","legendgroup":"(12hr,2)","showlegend":false,"xaxis":"x","yaxis":"y","hoverinfo":"text","frame":null}],"layout":{"margin":{"t":23.305936073059364,"r":7.3059360730593621,"b":37.260273972602747,"l":48.949771689497723},"plot_bgcolor":"rgba(235,235,235,1)","paper_bgcolor":"rgba(255,255,255,1)","font":{"color":"rgba(0,0,0,1)","family":"","size":14.611872146118724},"xaxis":{"domain":[0,1],"automargin":true,"type":"linear","autorange":false,"range":[0.29687565576217811,0.65291142899425969],"tickmode":"array","ticktext":["0.3","0.4","0.5","0.6"],"tickvals":[0.30000000000000004,0.40000000000000002,0.5,0.60000000000000009],"categoryorder":"array","categoryarray":["0.3","0.4","0.5","0.6"],"nticks":null,"ticks":"outside","tickcolor":"rgba(51,51,51,1)","ticklen":3.6529680365296811,"tickwidth":0.66417600664176002,"showticklabels":true,"tickfont":{"color":"rgba(77,77,77,1)","family":"","size":11.68949771689498},"tickangle":-0,"showline":false,"linecolor":null,"linewidth":0,"showgrid":true,"gridcolor":"rgba(255,255,255,1)","gridwidth":0.66417600664176002,"zeroline":false,"anchor":"y","title":{"text":"PC1","font":{"color":"rgba(0,0,0,1)","family":"","size":14.611872146118724}},"hoverformat":".2f"},"yaxis":{"domain":[0,1],"automargin":true,"type":"linear","autorange":false,"range":[0.4339568812386127,0.62507789398913349],"tickmode":"array","ticktext":["0.45","0.50","0.55","0.60"],"tickvals":[0.45000000000000001,0.5,0.55000000000000004,0.60000000000000009],"categoryorder":"array","categoryarray":["0.45","0.50","0.55","0.60"],"nticks":null,"ticks":"outside","tickcolor":"rgba(51,51,51,1)","ticklen":3.6529680365296811,"tickwidth":0.66417600664176002,"showticklabels":true,"tickfont":{"color":"rgba(77,77,77,1)","family":"","size":11.68949771689498},"tickangle":-0,"showline":false,"linecolor":null,"linewidth":0,"showgrid":true,"gridcolor":"rgba(255,255,255,1)","gridwidth":0.66417600664176002,"zeroline":false,"anchor":"x","title":{"text":"PC2","font":{"color":"rgba(0,0,0,1)","family":"","size":14.611872146118724}},"hoverformat":".2f"},"shapes":[{"type":"rect","fillcolor":null,"line":{"color":null,"width":0,"linetype":[]},"yref":"paper","xref":"paper","x0":0,"x1":1,"y0":0,"y1":1}],"showlegend":true,"legend":{"bgcolor":"rgba(255,255,255,1)","bordercolor":"transparent","borderwidth":1.8897637795275593,"font":{"color":"rgba(0,0,0,1)","family":"","size":11.68949771689498},"title":{"text":"Time<br />Rep","font":{"color":"rgba(0,0,0,1)","family":"","size":14.611872146118724}}},"hovermode":"closest","barmode":"relative"},"config":{"doubleClick":"reset","modeBarButtonsToAdd":["hoverclosest","hovercompare"],"showSendToCloud":false},"source":"select","attrs":{"169e10c853c2":{"x":{},"y":{},"colour":{},"shape":{},"type":"scatter"},"169e1acfdc28":{"x":{},"y":{},"colour":{},"shape":{},"text":{}}},"cur_data":"169e10c853c2","visdat":{"169e10c853c2":["function (y) ","x"],"169e1acfdc28":["function (y) ","x"]},"highlight":{"on":"plotly_click","persistent":false,"dynamic":false,"selectize":false,"opacityDim":0.20000000000000001,"selected":{"opacity":1},"debounce":0},"shinyEvents":["plotly_hover","plotly_click","plotly_selected","plotly_relayout","plotly_brushed","plotly_brushing","plotly_clickannotation","plotly_doubleclick","plotly_deselect","plotly_afterplot","plotly_sunburstclick"],"base_url":"https://plot.ly"},"evals":[],"jsHooks":[]}</script> --- Tables - DT Simlar to plots we can create interactive tables that are both searchable and sortable. We do this with the DT package. Lets make some quick demo data. ``` r label <- c("Gene1", "Gene2", "Gene3") temp <- rnorm(3) temp2 <- rnorm(3) dfExample <- data.frame(label, temp, temp2) dfExample ``` ``` ## label temp temp2 ## 1 Gene1 0.9020976 -2.4004368 ## 2 Gene2 -0.6781647 0.6485534 ## 3 Gene3 -0.6067576 -0.8492661 ``` --- DT We can simply use the datatable function to then create our interactive table. ``` r library(DT) datatable(dfExample) ``` <div class="datatables html-widget html-fill-item" id="htmlwidget-11f659531e57758e70dd" style="width:100%;height:auto;"></div> <script type="application/json" data-for="htmlwidget-11f659531e57758e70dd">{"x":{"filter":"none","vertical":false,"data":[["1","2","3"],["Gene1","Gene2","Gene3"],[0.9020975849815475,-0.6781646655083075,-0.6067575773900311],[-2.400436805901972,0.6485534079065669,-0.8492661250100386]],"container":"<table class=\"display\">\n <thead>\n <tr>\n <th> <\/th>\n <th>label<\/th>\n <th>temp<\/th>\n <th>temp2<\/th>\n <\/tr>\n <\/thead>\n<\/table>","options":{"columnDefs":[{"className":"dt-right","targets":[2,3]},{"orderable":false,"targets":0},{"name":" ","targets":0},{"name":"label","targets":1},{"name":"temp","targets":2},{"name":"temp2","targets":3}],"order":[],"autoWidth":false,"orderClasses":false},"selection":{"mode":"multiple","selected":null,"target":"row","selectable":null}},"evals":[],"jsHooks":[]}</script> --- DT There are many ways to [customize](https://rstudio.github.io/DT/options.html) our table i.e. removing rownames, adding titles, default sorting etc ``` r datatable(dfExample, rownames = FALSE, caption = "My Table") ``` <div class="datatables html-widget html-fill-item" id="htmlwidget-81826ce318555014aba2" style="width:100%;height:auto;"></div> <script type="application/json" data-for="htmlwidget-81826ce318555014aba2">{"x":{"filter":"none","vertical":false,"caption":"<caption>My Table<\/caption>","data":[["Gene1","Gene2","Gene3"],[0.9020975849815475,-0.6781646655083075,-0.6067575773900311],[-2.400436805901972,0.6485534079065669,-0.8492661250100386]],"container":"<table class=\"display\">\n <thead>\n <tr>\n <th>label<\/th>\n <th>temp<\/th>\n <th>temp2<\/th>\n <\/tr>\n <\/thead>\n<\/table>","options":{"columnDefs":[{"className":"dt-right","targets":[1,2]},{"name":"label","targets":0},{"name":"temp","targets":1},{"name":"temp2","targets":2}],"order":[],"autoWidth":false,"orderClasses":false},"selection":{"mode":"multiple","selected":null,"target":"row","selectable":null}},"evals":[],"jsHooks":[]}</script> --- class: inverse, center, middle # Quarto <html><div style='float:left'></div><hr color='#EB811B' size=1px width=720px></html> --- # Quarto Rmarkdown has been an incredibly useful tool for building reports for a long time, but it is predominately focused on R (though it can be used indirectly for python). We have also mentioned [Jupyter Notebook](https://jupyter.org/). It is a similar notebook system. It was built for Python (though it can be used indirectly for R). [Quarto](https://quarto.org/) is a new form of report built on Rmds that can handle python and other languages in a more native way. --- # Rmarkdown and Quarto Quarto is supported by Posit, the people behind RStudio (who also developed Rmarkdown). Right now if you work in R, Rmarkdown will suffice, but the use of Quarto is growing and ultimately it will be the succesor. Luckily everything you have just learnt about Rmd formatting will work for Quarto Markdowns (qmds) too. --- # Why Quarto? It will natively allow you to run code for: R, Python, Julia, JavaScript. Bioinformatics has a rich history with R, while Python has a wealth of machine learning packages. With the more recent developments in Deep leanring and AI more folks are dabbling with specific Python tools for parts of their analysis. As a result bioinformatics is becoming increasingly multi-language. --- # Why Quarto? There is a lot of active development to add new features not found in Rmds: * Quarto is not dependent on R or RStudio * More [Output Formats](https://quarto.org/docs/output-formats/all-formats.html) - Native Reveal.js or powerpoint slides * Enhanced [interacivity] - Can be done locally with [Observable JavScript](https://quarto.org/docs/computations/ojs.html) or hooked into servers to run dashboards through [Shiny](https://quarto.org/docs/dashboards/interactivity/) * Better [template](https://quarto.org/docs/extensions/starter-templates.html) creation * Built-in [citation management](https://quarto.org/docs/authoring/citations.html) & bibliography formatting * Broadly increased [customization](https://quarto.org/docs/output-formats/page-layout.html) --- # How to make a Qmd? In Rstudio it is the same as making a Rmd. Just pick a different option: New > Quarto Document This should largely look the same. --- # Quarto Code Chunks .pull-left[ **Quarto** ```` default ```{r} #| warning=F #| message=F library(ggplot2) ``` ```` ] .pull-right[ **Rmarkdown** ```` default ```{r,warning=F,message=F} library(ggplot2) ``` ```` This will be accepted by Quarto regardless. There is backwards compatability. ] --- # Quarto YAML Most of the YAML will work in the same way, though there are slight differenes in the preferred names. .pull-left[ **Quarto** ``` default --- format: html: fig-width: 7 fig-height: 6 --- ``` ] .pull-right[ **Rmarkdown** ``` default --- output: html_document: fig_width: 7 fig_height: 6 --- ``` This will be accepted by Quarto regardless. There is some backwards compatability. ] --- # A key difference Resource like images are not embedded by default into your report when you are using Quarto. This means if you move your report to share it, unless you also move the files associated with the rendered html i.e. MyReport.html and MyReport_files. To ensure everything you need is embedded into the final document you just need to add *embed-resources: true* to the YAML. ``` default --- format: html: embed-resources: true fig-width: 7 fig-height: 6 --- ``` --- # An example YAML ``` default --- title: "My_Project_Name" author: - "Matt Paul" - "Bioinformatics Resource Center" - "Rockefeller University" date: "`r format(Sys.Date(), '%m/%d/%Y')`" format: html: embed-resources: true code_folding: hide number_sections: true theme: yeti highlight: tango toc: true toc_float: true --- ``` --- # Quarto and Jupyter As mentioned many people who already are making reports, use Jupyter. These are typically *.ipynb* files. Quarto can be used to render these files directly. To do this you just need to add a new cell at the top containing our YAML. Code arguments can also be added. <div align="center"> <img src="imgs/jupyter.png" alt="jupyter" height="300" width="500"> </div> --- # Quarto and Jupyter You just need to run Quarto directly to render the Jupyter notebook. You will run this opn the command line. ``` sh quarto render notebook.ipynb --execute ``` 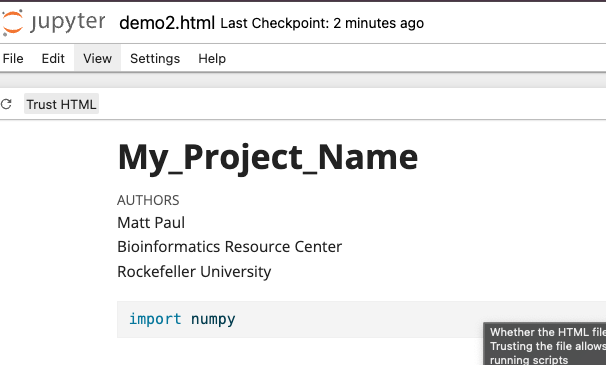 --- # Quarto and VS Code We have mostly been showing you how to use Quarto with RStudio. But if you are a VS Code user you can just install the Quarto extension there and use it within VS Code. --- # Quarto and the future Rmarkdown and Jupyter are great and widely supported. They will likely be superseded by Quarto over time. To summarize: * Rmarkdown and Jupyter will largely work with Quarto already * New features and increased customization option * You will probably want to use *embed-resources* --- ## Resources * [Rmarkdown website](http://rmarkdown.rstudio.com/) * [Rmarkdown Book](https://bookdown.org/yihui/rmarkdown/) * [Rmarkdown Cheatsheet Download](https://raw.githubusercontent.com/rstudio/cheatsheets/main/rmarkdown.pdf) * [Quarto](https://quarto.org/) --- ## Exercises Exercise on Reproducibility Reports can be found [here](../../exercises/exercises/ReproducibleR_exercise.html) --- ## Contact Any suggestions, comments, edits or questions (about content or the slides themselves) please reach out to our [GitHub](https://github.com/RockefellerUniversity/Reproducible_R/issues) and raise an issue.